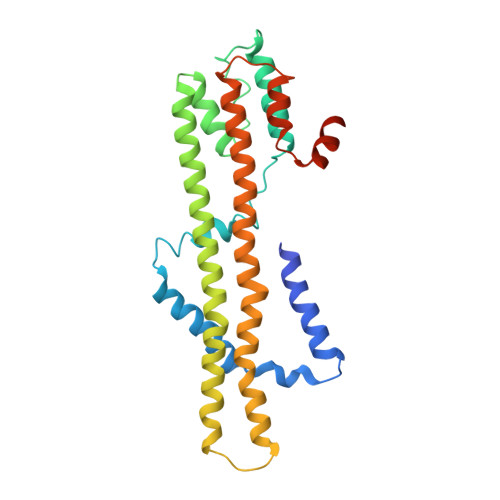
Structures of the stator complex that drives rotation of the bacterial flagellum.
Deme, J.C., Johnson, S., Vickery, O., Aron, A., Monkhouse, H., Griffiths, T., James, R.H., Berks, B.C., Coulton, J.W., Stansfeld, P.J., Lea, S.M.(2020) Nat Microbiol 5: 1553-1564
- PubMed: 32929189
- DOI: https://doi.org/10.1038/s41564-020-0788-8
- Primary Citation of Related Structures:
6YSF, 6YSL - PubMed Abstract:
The bacterial flagellum is the prototypical protein nanomachine and comprises a rotating helical propeller attached to a membrane-embedded motor complex. The motor consists of a central rotor surrounded by stator units that couple ion flow across the cytoplasmic membrane to generate torque. Here, we present the structures of the stator complexes from Clostridium sporogenes, Bacillus subtilis and Vibrio mimicus, allowing interpretation of the extensive body of data on stator mechanism. The structures reveal an unexpected asymmetric A 5 B 2 subunit assembly where the five A subunits enclose the two B subunits. Comparison to structures of other ion-driven motors indicates that this A 5 B 2 architecture is fundamental to bacterial systems that couple energy from ion flow to generate mechanical work at a distance and suggests that such events involve rotation in the motor structures.
Organizational Affiliation:
Sir William Dunn School of Pathology, University of Oxford, Oxford, UK.