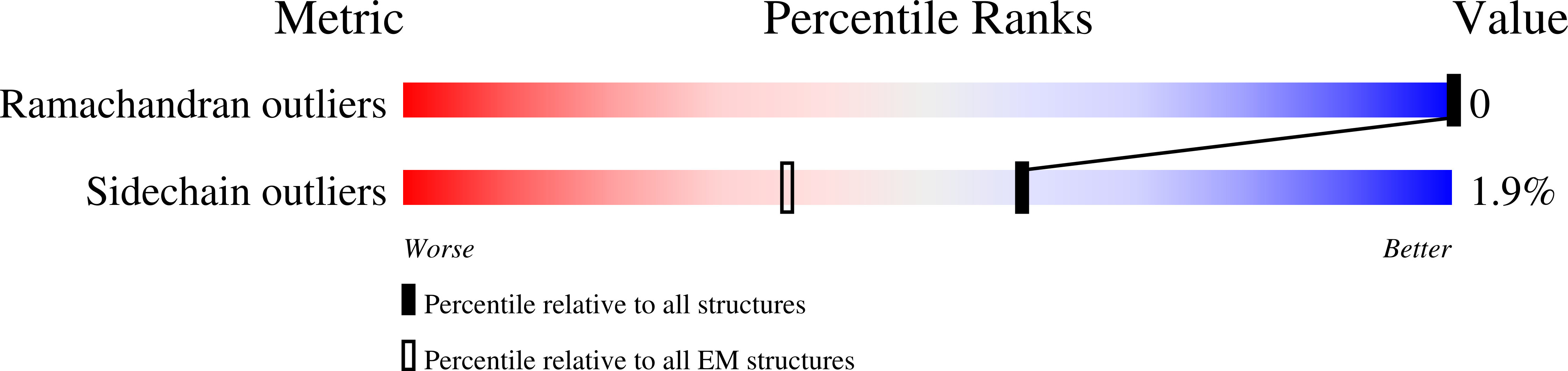High-Light versus Low-Light: Effects on Paired Photosystem II Supercomplex Structural Rearrangement in Pea Plants.
Grinzato, A., Albanese, P., Marotta, R., Swuec, P., Saracco, G., Bolognesi, M., Zanotti, G., Pagliano, C.(2020) Int J Mol Sci 21
- PubMed: 33207833
- DOI: https://doi.org/10.3390/ijms21228643
- Primary Citation of Related Structures:
6YP7 - PubMed Abstract:
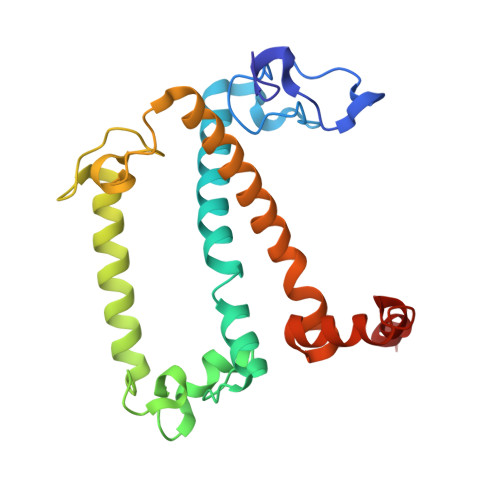
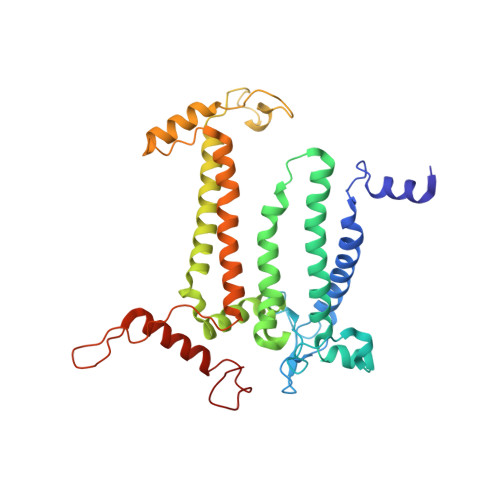
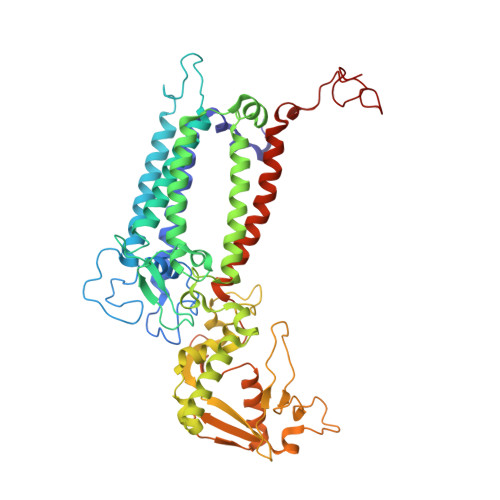
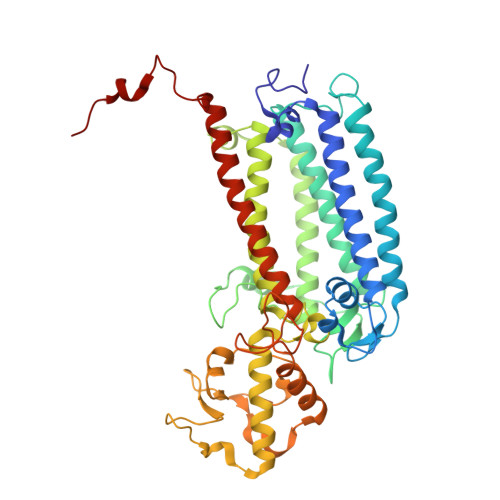
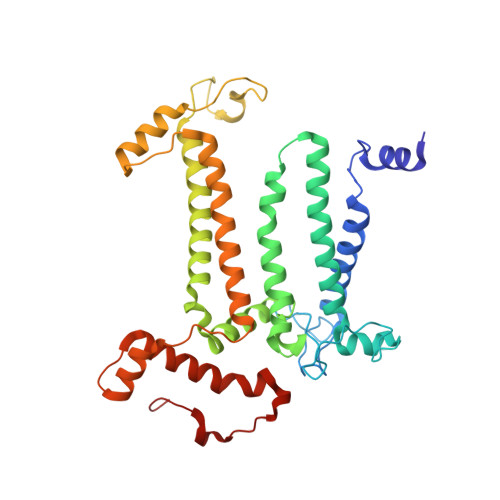
In plant grana thylakoid membranes Photosystem II (PSII) associates with a variable number of antenna proteins (LHCII) to form different types of supercomplexes (PSII-LHCII), whose organization is dynamically adjusted in response to light cues, with the C 2 S 2 more abundant in high-light and the C 2 S 2 M 2 in low-light. Paired PSII-LHCII supercomplexes interacting at their stromal surface from adjacent thylakoid membranes were previously suggested to mediate grana stacking. Here, we present the cryo-electron microscopy maps of paired C 2 S 2 and C 2 S 2 M 2 supercomplexes isolated from pea plants grown in high-light and low-light, respectively. These maps show a different rotational offset between the two supercomplexes in the pair, responsible for modifying their reciprocal interaction and energetic connectivity. This evidence reveals a different way by which paired PSII-LHCII supercomplexes can mediate grana stacking at diverse irradiances. Electrostatic stromal interactions between LHCII trimers almost completely overlapping in the paired C 2 S 2 can be the main determinant by which PSII-LHCII supercomplexes mediate grana stacking in plants grown in high-light, whereas the mutual interaction of stromal N-terminal loops of two facing Lhcb4 subunits in the paired C 2 S 2 M 2 can fulfil this task in plants grown in low-light. The high-light induced accumulation of the Lhcb4.3 protein in PSII-LHCII supercomplexes has been previously reported. Our cryo-electron microscopy map at 3.8 Å resolution of the C 2 S 2 supercomplex isolated from plants grown in high-light suggests the presence of the Lhcb4.3 protein revealing peculiar structural features of this high-light-specific antenna important for photoprotection.
Organizational Affiliation:
Department of Biomedical Sciences, University of Padova, Via Ugo Bassi 58 B, 35121 Padova, Italy.