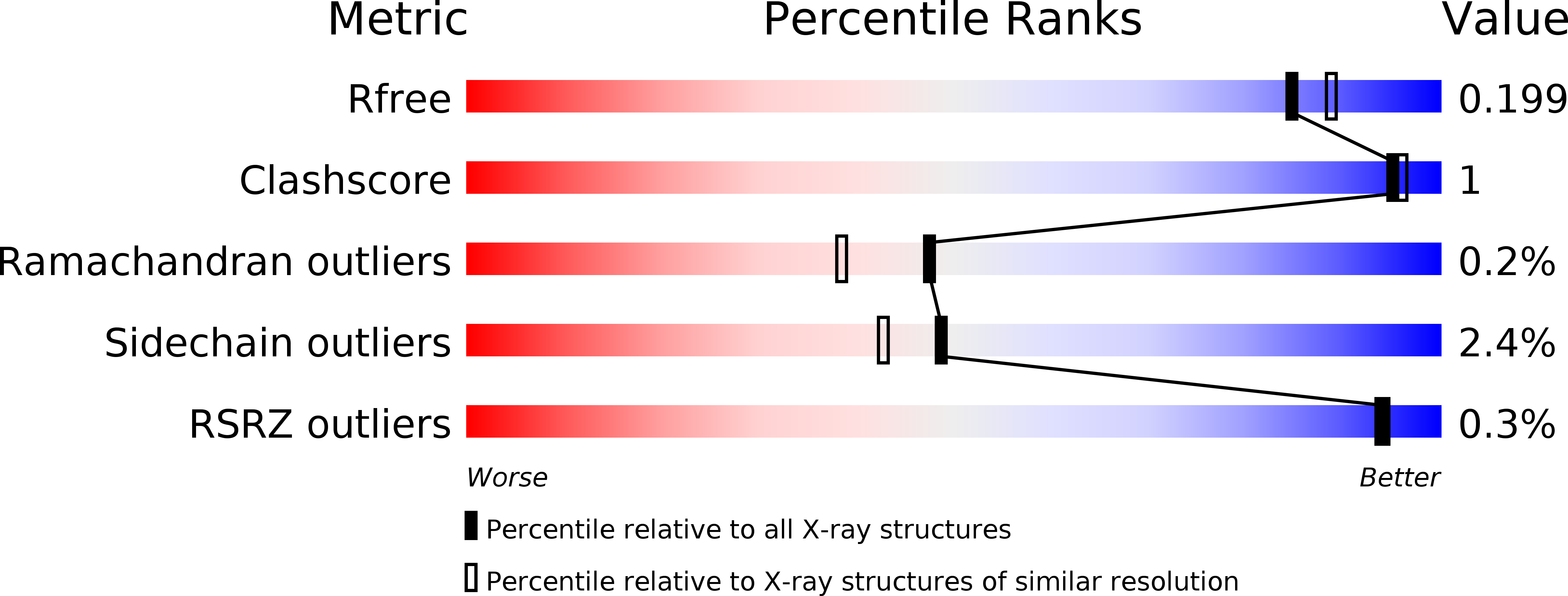
The co-existence of cold activity and thermal stability in an Antarctic GH42 beta-galactosidase relies on its hexameric quaternary arrangement.
Mangiagalli, M., Lapi, M., Maione, S., Orlando, M., Brocca, S., Pesce, A., Barbiroli, A., Camilloni, C., Pucciarelli, S., Lotti, M., Nardini, M.(2021) FEBS J 288: 546-565
- PubMed: 32363751
- DOI: https://doi.org/10.1111/febs.15354
- Primary Citation of Related Structures:
6Y2K - PubMed Abstract:
To survive in cold environments, psychrophilic organisms produce enzymes endowed with high specific activity at low temperature. The structure of these enzymes is usually flexible and mostly thermolabile. In this work, we investigate the structural basis of cold adaptation of a GH42 β-galactosidase from the psychrophilic Marinomonas ef1. This enzyme couples cold activity with astonishing robustness for a psychrophilic protein, for it retains 23% of its highest activity at 5 °C and it is stable for several days at 37 °C and even 50 °C. Phylogenetic analyses indicate a close relationship with thermophilic β-galactosidases, suggesting that the present-day enzyme evolved from a thermostable scaffold modeled by environmental selective pressure. The crystallographic structure reveals the overall similarity with GH42 enzymes, along with a hexameric arrangement (dimer of trimers) not found in psychrophilic, mesophilic, and thermophilic homologues. In the quaternary structure, protomers form a large central cavity, whose accessibility to the substrate is promoted by the dynamic behavior of surface loops, even at low temperature. A peculiar cooperative behavior of the enzyme is likely related to the increase of the internal cavity permeability triggered by heating. Overall, our results highlight a novel strategy of enzyme cold adaptation, based on the oligomerization state of the enzyme, which effectively challenges the paradigm of cold activity coupled with intrinsic thermolability. DATABASE: Structural data are available in the Protein Data Bank database under the accession number 6Y2K.
Organizational Affiliation:
Department of Biotechnology and Biosciences, University of Milano-Bicocca, Italy.