A new perspective on the interaction between the Vg/VGLL1-3 proteins and the TEAD transcription factors.
Mesrouze, Y., Aguilar, G., Bokhovchuk, F., Martin, T., Delaunay, C., Villard, F., Meyerhofer, M., Zimmermann, C., Fontana, P., Wille, R., Vorherr, T., Erdmann, D., Furet, P., Scheufler, C., Schmelzle, T., Affolter, M., Chene, P.(2020) Sci Rep 10: 17442-17442
- PubMed: 33060790
- DOI: https://doi.org/10.1038/s41598-020-74584-x
- Primary Citation of Related Structures:
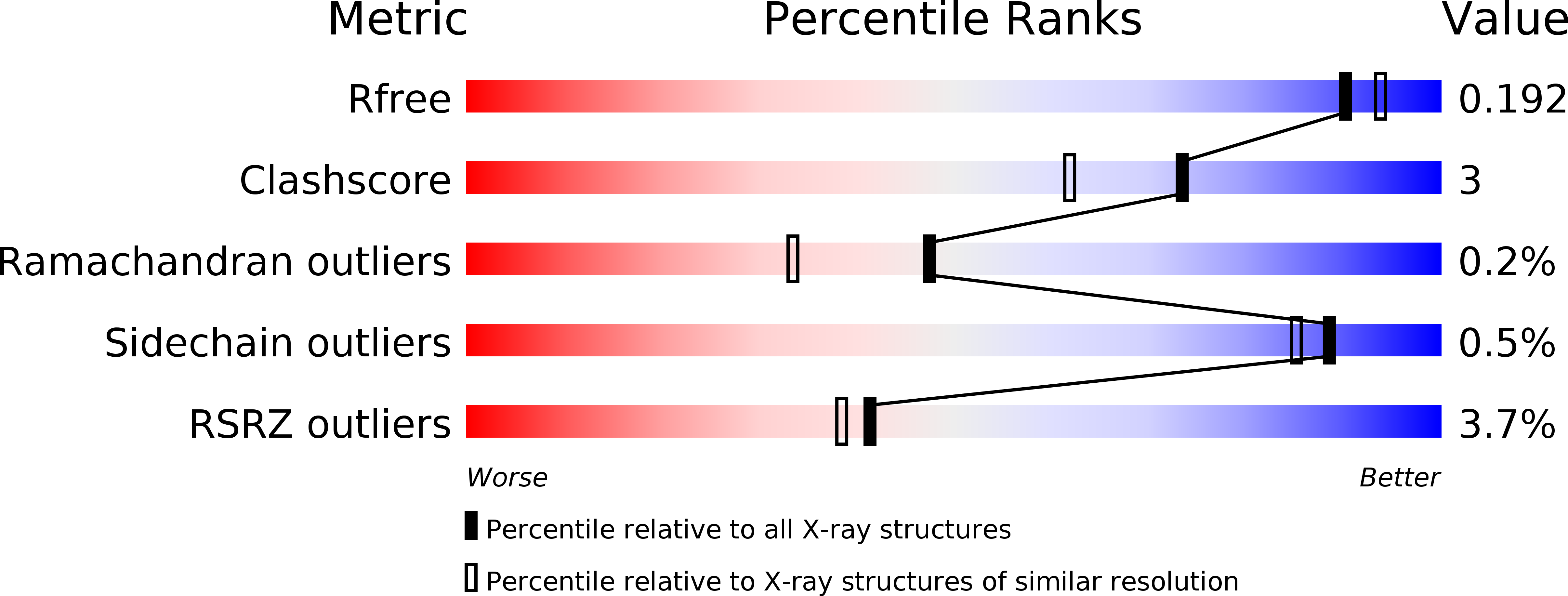
6Y20 - PubMed Abstract:
The most downstream elements of the Hippo pathway, the TEAD transcription factors, are regulated by several cofactors, such as Vg/VGLL1-3. Earlier findings on human VGLL1 and here on human VGLL3 show that these proteins interact with TEAD via a conserved amino acid motif called the TONDU domain. Surprisingly, our studies reveal that the TEAD-binding domain of Drosophila Vg and of human VGLL2 is more complex and contains an additional structural element, an Ω-loop, that contributes to TEAD binding. To explain this unexpected structural difference between proteins from the same family, we propose that, after the genome-wide duplications at the origin of vertebrates, the Ω-loop present in an ancestral VGLL gene has been lost in some VGLL variants. These findings illustrate how structural and functional constraints can guide the evolution of transcriptional cofactors to preserve their ability to compete with other cofactors for binding to transcription factors.
Organizational Affiliation:
Disease Area Oncology, Novartis Institutes for Biomedical Research, Novartis, WSJ 386 4.02.01, 4002, Basel, Switzerland.