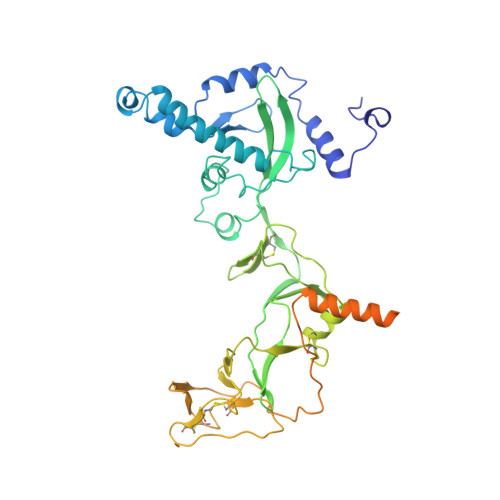
Cryo-EM structure of the prefusion state of canine distemper virus fusion protein ectodomain.
Kalbermatter, D., Shrestha, N., Gall, F.M., Wyss, M., Riedl, R., Plattet, P., Fotiadis, D.(2020) J Struct Biol X 4: 100021-100021
- PubMed: 32647825
- DOI: https://doi.org/10.1016/j.yjsbx.2020.100021
- Primary Citation of Related Structures:
6XYE - PubMed Abstract:
Measles virus (MeV) and canine distemper virus (CDV), two members of the Morbillivirus genus, are still causing important global diseases of humans and animals, respectively. To enter target cells, morbilliviruses rely on an envelope-anchored machinery, which is composed of two interacting glycoproteins: a tetrameric receptor binding (H) protein and a trimeric fusion (F) protein. To execute membrane fusion, the F protein initially adopts a metastable, prefusion state that refolds into a highly stable postfusion conformation as the result of a finely coordinated activation process mediated by the H protein. Here, we employed cryo-electron microscopy (cryo-EM) and single particle reconstruction to elucidate the structure of the prefusion state of the CDV F protein ectodomain (solF) at 4.3 Å resolution. Stabilization of the prefusion solF trimer was achieved by fusing the GCNt trimerization sequence at the C-terminal protein region, and expressing and purifying the recombinant protein in the presence of a morbilliviral fusion inhibitor class compound. The three-dimensional cryo-EM map of prefusion CDV solF in complex with the inhibitor clearly shows density for the ligand at the protein binding site suggesting common mechanisms of membrane fusion activation and inhibition employed by different morbillivirus members.
Organizational Affiliation:
Institute of Biochemistry and Molecular Medicine, University of Bern, Bern, Switzerland.