A Key Motif in the Cholesterol-Dependent Cytolysins Reveals a Large Family of Related Proteins.
Evans, J.C., Johnstone, B.A., Lawrence, S.L., Morton, C.J., Christie, M.P., Parker, M.W., Tweten, R.K.(2020) mBio 11
- PubMed: 32994330
- DOI: https://doi.org/10.1128/mBio.02351-20
- Primary Citation of Related Structures:
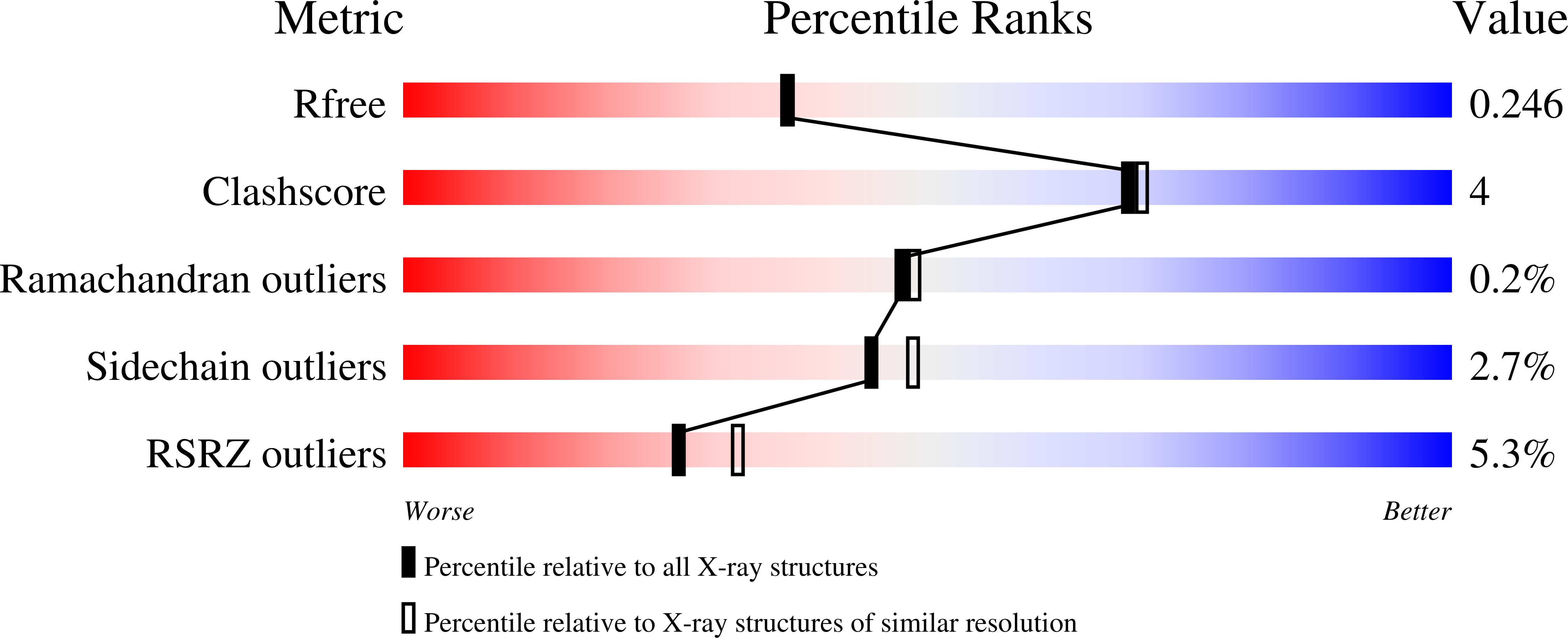
6XD4 - PubMed Abstract:
The cholesterol-dependent cytolysins (CDCs) are bacterial, β-barrel, pore-forming toxins. A central enigma of the pore-forming mechanism is how completion of the prepore is sensed to initiate its conversion to the pore. We identified a motif that is conserved between the CDCs and a diverse family of nearly 300 uncharacterized proteins present in over 220 species that span at least 10 bacterial and 2 eukaryotic phyla. Except for this motif, these proteins exhibit little similarity to the CDCs at the primary structure level. Studies herein show this motif is a critical component of the sensor that initiates the prepore-to-pore transition in the CDCs. We further show by crystallography, single particle analysis, and biochemical studies of one of these CDC-like (CDCL) proteins from Elizabethkingia anophelis , a commensal of the malarial mosquito midgut, that a high degree of structural similarity exists between the CDC and CDCL monomer structures and both form large oligomeric pore complexes. Furthermore, the conserved motif in the E. anophelis CDCL crystal structure occupies a nearly identical position and makes similar contacts to those observed in the structure of the archetype CDC, perfringolysin O (PFO). This suggests a common function in the CDCs and CDCLs and may explain why only this motif is conserved in the CDCLs. Hence, these studies identify a critical component of the sensor involved in initiating the prepore-to-pore transition in the CDCs, which is conserved in a large and diverse group of distant relatives of the CDCs. IMPORTANCE The cholesterol-dependent cytolysins' pore-forming mechanism relies on the ability to sense the completion of the oligomeric prepore structure and initiate the insertion of the β-barrel pore from the assembled prepore structure. These studies show that a conserved motif is an important component of the sensor that triggers the prepore-to-pore transition and that it is conserved in a large family of previously unidentified CDC-like proteins, the genes for which are present in a vast array of microbial species that span most terrestrial environments, as well as most animal and human microbiomes. These studies establish the foundation for future investigations that will probe the contribution of this large family of CDC-like proteins to microbial survival and human disease.
Organizational Affiliation:
Department of Microbiology and Immunology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma, USA.