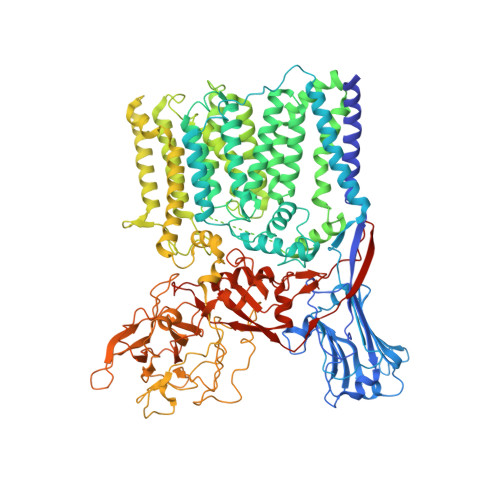
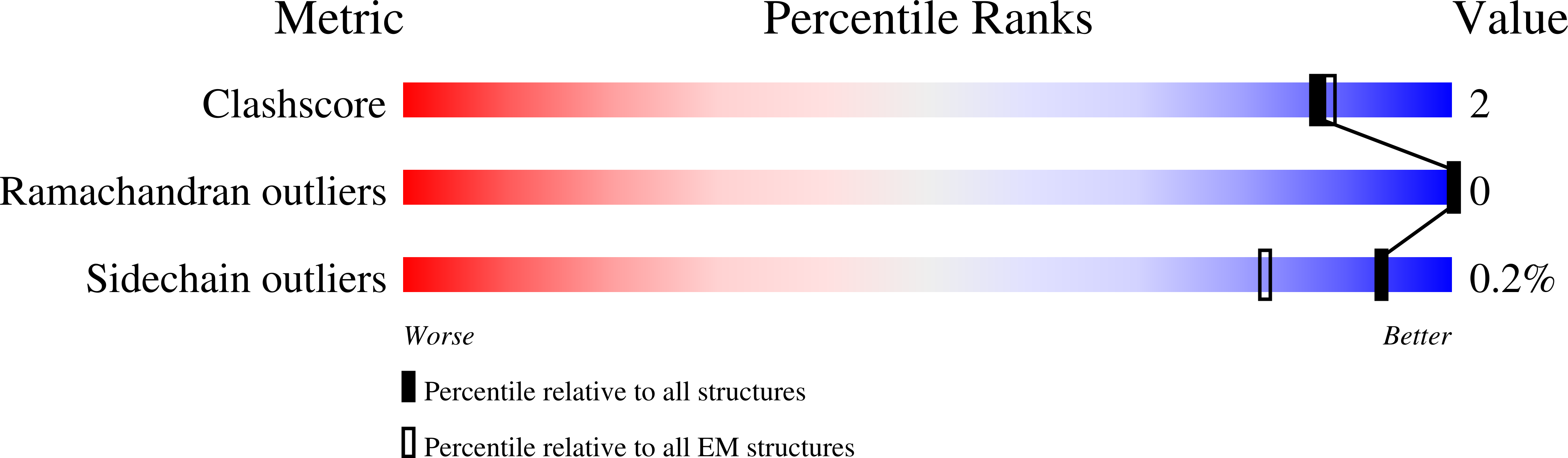Cryo-EM structure of arabinosyltransferase EmbB from Mycobacterium smegmatis.
Tan, Y.Z., Rodrigues, J., Keener, J.E., Zheng, R.B., Brunton, R., Kloss, B., Giacometti, S.I., Rosario, A.L., Zhang, L., Niederweis, M., Clarke, O.B., Lowary, T.L., Marty, M.T., Archer, M., Potter, C.S., Carragher, B., Mancia, F.(2020) Nat Commun 11: 3396-3396
- PubMed: 32636380
- DOI: https://doi.org/10.1038/s41467-020-17202-8
- Primary Citation of Related Structures:
6X0O - PubMed Abstract:
Arabinosyltransferase B (EmbB) belongs to a family of membrane-bound glycosyltransferases that build the lipidated polysaccharides of the mycobacterial cell envelope, and are targets of anti-tuberculosis drug ethambutol. We present the 3.3 Å resolution single-particle cryo-electron microscopy structure of Mycobacterium smegmatis EmbB, providing insights on substrate binding and reaction mechanism. Mutations that confer ethambutol resistance map mostly around the putative active site, suggesting this to be the location of drug binding.
Organizational Affiliation:
Department of Physiology and Cellular Biophysics, Columbia University, New York, NY, 10032, USA.