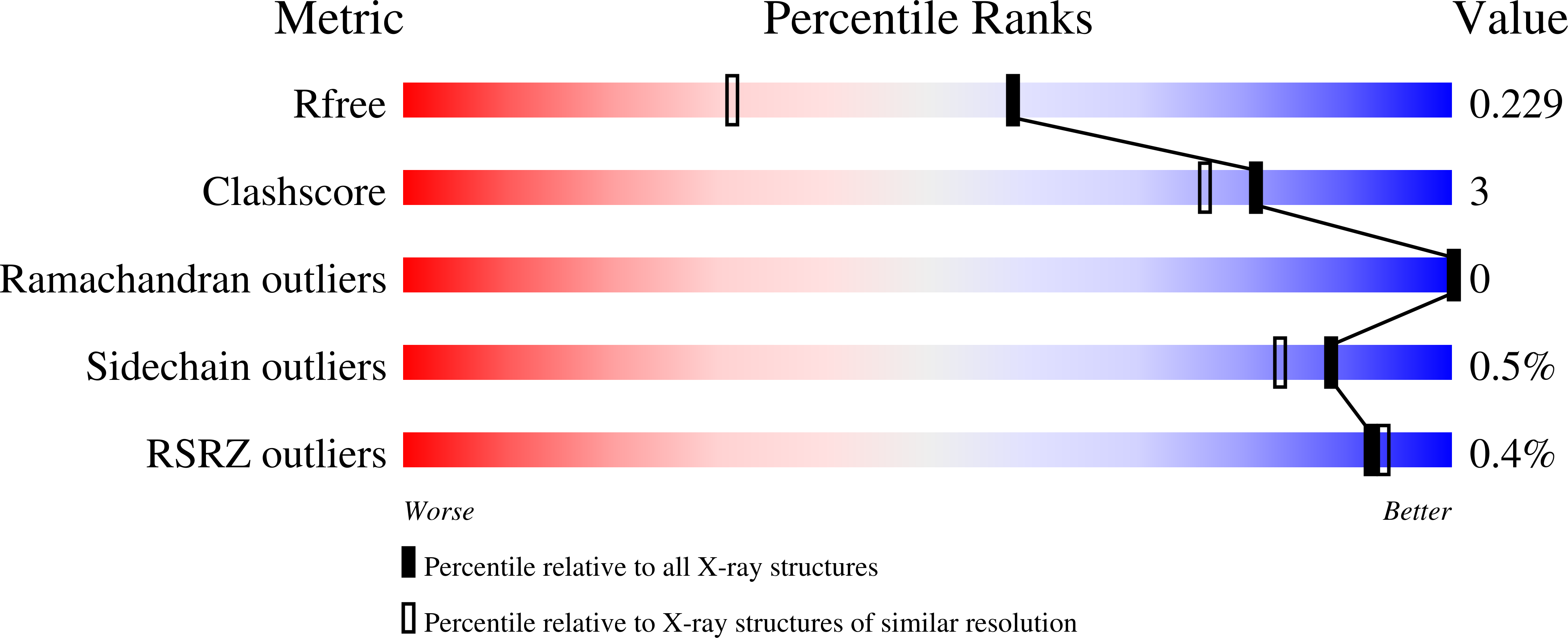
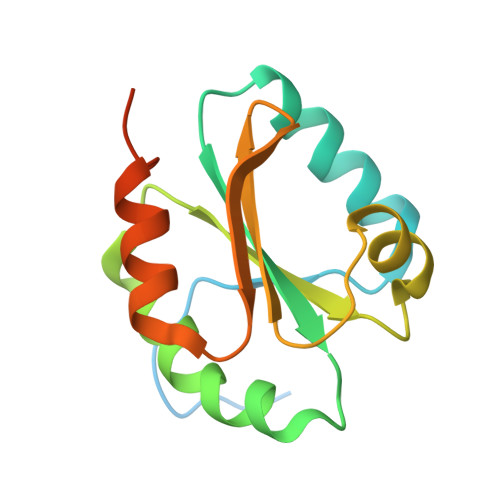
High resolution crystal structure of NaTrxh from Nicotiana alata and its interaction with the S-RNase.
Torres-Rodriguez, M.D., Gonzalez-Segura, L., Rodriguez-Sotres, R., Juarez-DiaZ, J.A., Cruz-Zamora, Y., Cruz-Garcia, F.(2020) J Struct Biol 212: 107578-107578
- PubMed: 32682729
- DOI: https://doi.org/10.1016/j.jsb.2020.107578
- Primary Citation of Related Structures:
6X0B - PubMed Abstract:
Thioredoxins are regulatory proteins that reduce disulfide bonds on target proteins. NaTrxh, which belongs to the plant thioredoxin family h subgroup 2, interacts and reduces the S-RNase enhancing its ribonuclease activity seven-fold, resulting an essential protein for pollen rejection inNicotiana.Here, the crystal structure of NaTrxh at 1.7 Å by X-ray diffraction is reported. NaTrxh conserves the typical fold observed in other thioredoxins from prokaryotes and eukaryotes, but it contains extensions towards both N- and C-termini.The NaTrxh N-terminal extension participates in the reduction of S-RNase, and in the structure reported here, this is orientated towards the reactive site. The interaction between S F11 -RNase and the NaTrxh N-terminal was simulated and the short-lived complex observed lasted for a tenth of ns. Moreover, we identified certain amino acids as S F11 -RNase-E155 and NaTrxh-M104 as good candidates to contribute to the stability of the complex. Furthermore, we simulated the reduction of the C153-C186 S F11 -RNase disulfide bond and observed subtle changes that affect the entire core, which might explain the increase in the ribonuclease activity of S-RNase when it is reduced by NaTrxh.
Organizational Affiliation:
Departamento de Bioquímica, Facultad de Química, Universidad Nacional Autónoma de México, Cd. Mx 04510, Mexico.