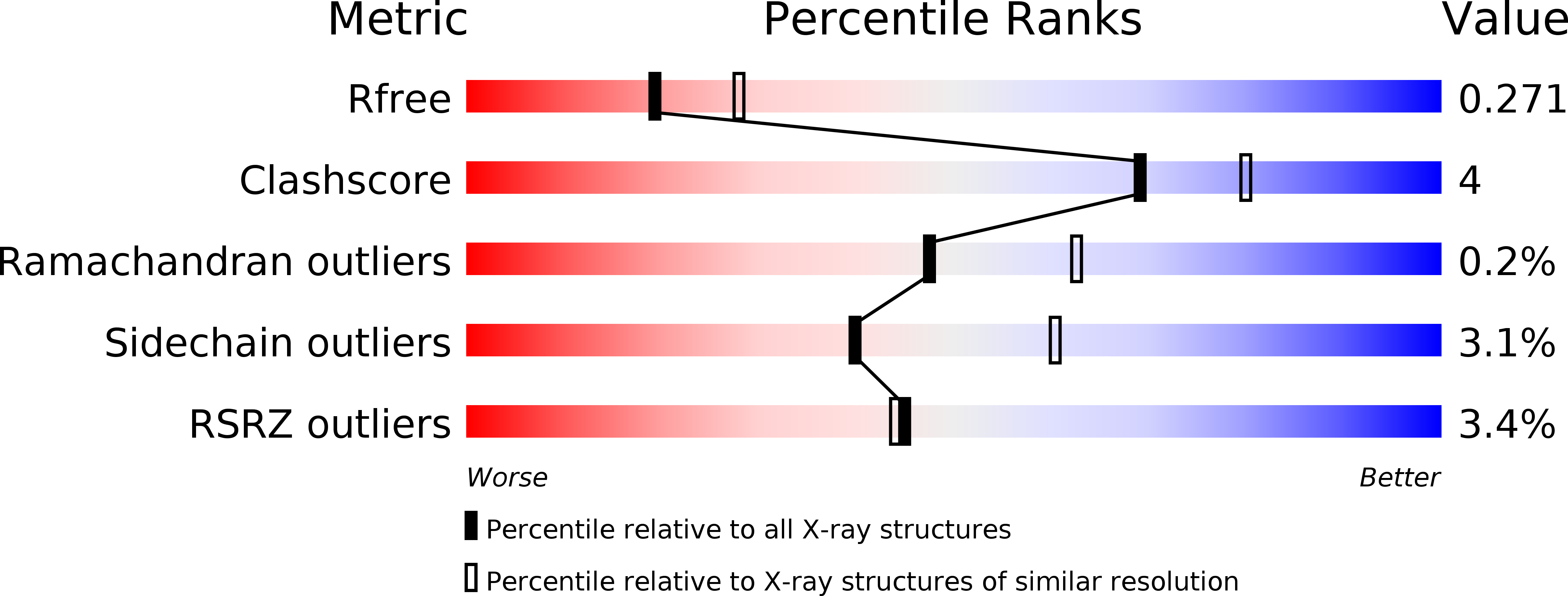
Biochemical basis for the regulation of biosynthesis of antiparasitics by bacterial hormones.
Kapoor, I., Olivares, P., Nair, S.K.(2020) Elife 9
- PubMed: 32510324
- DOI: https://doi.org/10.7554/eLife.57824
- Primary Citation of Related Structures:
6WP7, 6WP9, 6WPA - PubMed Abstract:
Diffusible small molecule microbial hormones drastically alter the expression profiles of antibiotics and other drugs in actinobacteria. For example, avenolide (a butenolide) regulates the production of avermectin, derivatives of which are used in the treatment of river blindness and other parasitic diseases. Butenolides and γ-butyrolactones control the production of pharmaceutically important secondary metabolites by binding to TetR family transcriptional repressors. Here, we describe a concise, 22-step synthetic strategy for the production of avenolide. We present crystal structures of the butenolide receptor AvaR1 in isolation and in complex with avenolide, as well as those of AvaR1 bound to an oligonucleotide derived from its operator. Biochemical studies guided by the co-crystal structures enable the identification of 90 new actinobacteria that may be regulated by butenolides, two of which are experimentally verified. These studies provide a foundation for understanding the regulation of microbial secondary metabolite production, which may be exploited for the discovery and production of novel medicines.
Organizational Affiliation:
Department of Biochemistry, University of Illinois at Urbana Champaign, Urbana, United States.