ZNF410 Uniquely Activates the NuRD Component CHD4 to Silence Fetal Hemoglobin Expression.
Lan, X., Ren, R., Feng, R., Ly, L.C., Lan, Y., Zhang, Z., Aboreden, N., Qin, K., Horton, J.R., Grevet, J.D., Mayuranathan, T., Abdulmalik, O., Keller, C.A., Giardine, B., Hardison, R.C., Crossley, M., Weiss, M.J., Cheng, X., Shi, J., Blobel, G.A.(2021) Mol Cell 81: 239-254.e8
- PubMed: 33301730
- DOI: https://doi.org/10.1016/j.molcel.2020.11.006
- Primary Citation of Related Structures:
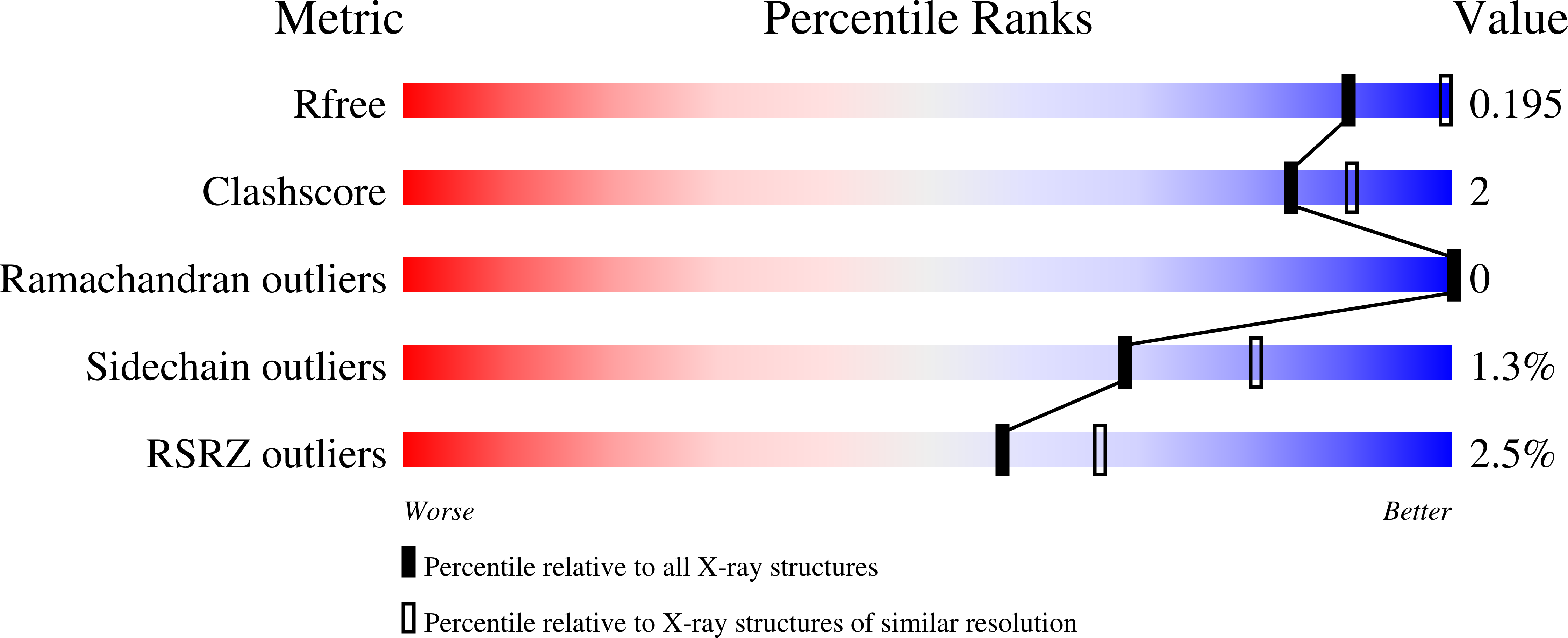
6WMI - PubMed Abstract:
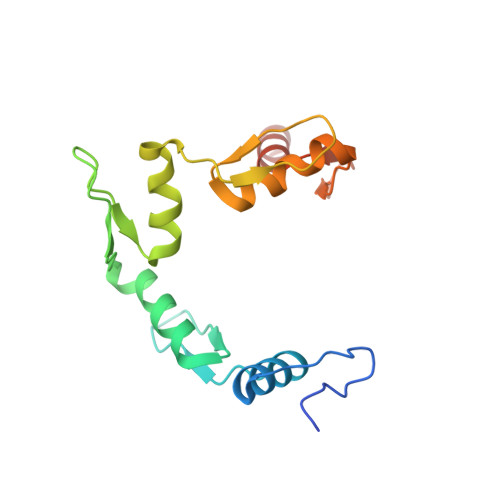
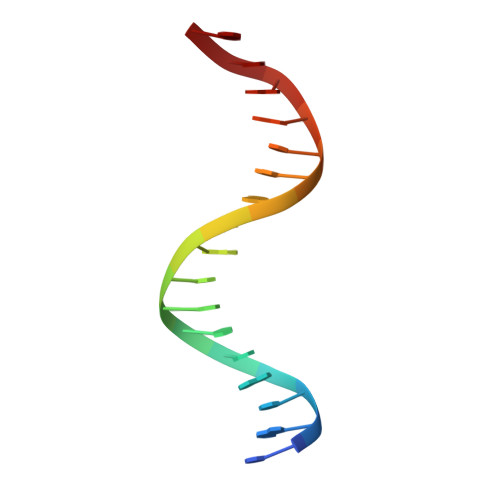
Metazoan transcription factors typically regulate large numbers of genes. Here we identify via a CRISPR-Cas9 genetic screen ZNF410, a pentadactyl DNA-binding protein that in human erythroid cells directly activates only a single gene, the NuRD component CHD4. Specificity is conveyed by two highly evolutionarily conserved clusters of ZNF410 binding sites near the CHD4 gene with no counterparts elsewhere in the genome. Loss of ZNF410 in adult-type human erythroid cell culture systems and xenotransplantation settings diminishes CHD4 levels and derepresses the fetal hemoglobin genes. While previously known to be silenced by CHD4, the fetal globin genes are exposed here as among the most sensitive to reduced CHD4 levels.. In vitro DNA binding assays and crystallographic studies reveal the ZNF410-DNA binding mode. ZNF410 is a remarkably selective transcriptional activator in erythroid cells, and its perturbation might offer new opportunities for treatment of hemoglobinopathies.
Organizational Affiliation:
Division of Hematology, The Children's Hospital of Philadelphia, Philadelphia, PA 19104, USA.