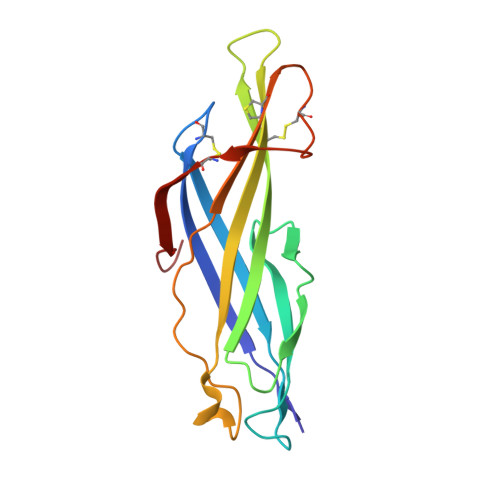
The crystal structure of SnTox3 from the necrotrophic fungus Parastagonospora nodorum reveals a unique effector fold and provides insight into Snn3 recognition and pro-domain protease processing of fungal effectors.
Outram, M.A., Sung, Y.C., Yu, D., Dagvadorj, B., Rima, S.A., Jones, D.A., Ericsson, D.J., Sperschneider, J., Solomon, P.S., Kobe, B., Williams, S.J.(2021) New Phytol 231: 2282-2296
- PubMed: 34053091
- DOI: https://doi.org/10.1111/nph.17516
- Primary Citation of Related Structures:
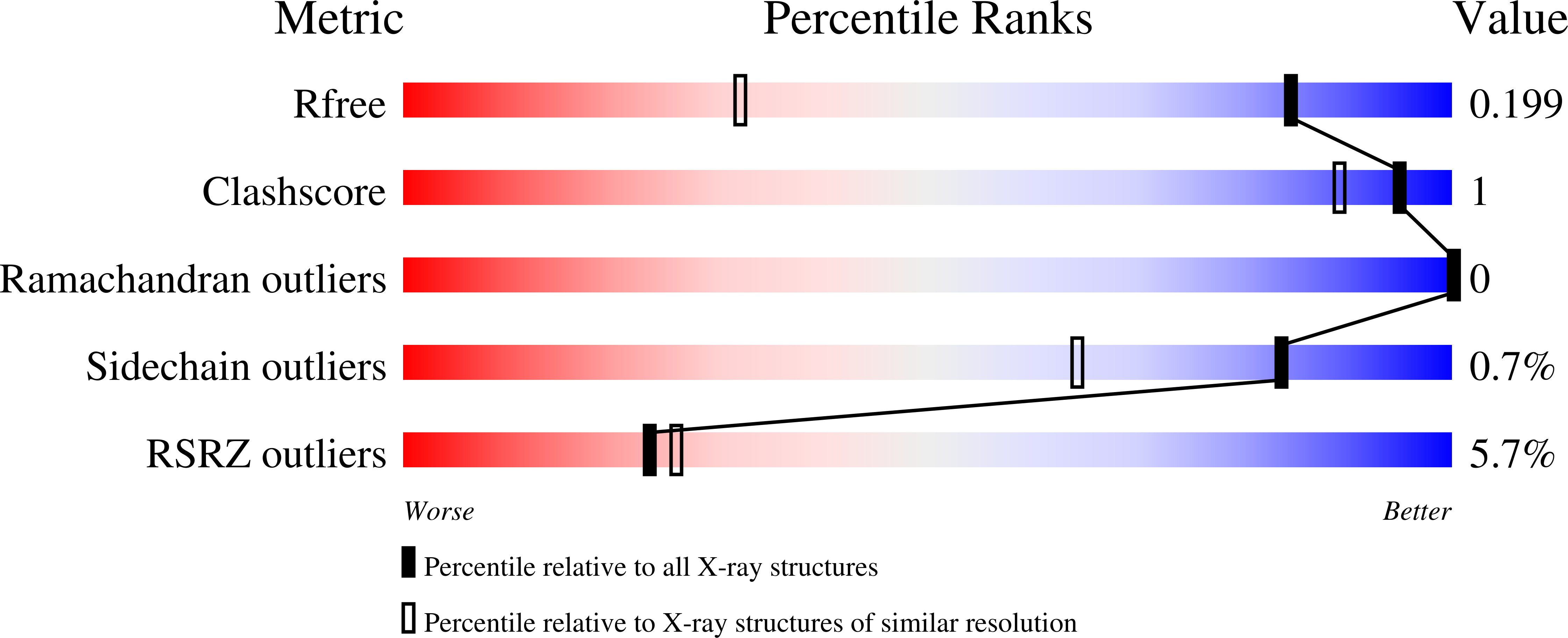
6WES - PubMed Abstract:
Plant pathogens cause disease through secreted effector proteins, which act to promote infection. Typically, the sequences of effectors provide little functional information and further targeted experimentation is required. Here, we utilized a structure/function approach to study SnTox3, an effector from the necrotrophic fungal pathogen Parastagonospora nodorum, which causes cell death in wheat-lines carrying the sensitivity gene Snn3. We developed a workflow for the production of SnTox3 in a heterologous host that enabled crystal structure determination and functional studies. We show this approach can be successfully applied to study effectors from other pathogenic fungi. The β-barrel fold of SnTox3 is a novel fold among fungal effectors. Structure-guided mutagenesis enabled the identification of residues required for Snn3 recognition. SnTox3 is a pre-pro-protein, and the pro-domain of SnTox3 can be cleaved in vitro by the protease Kex2. Complementing this, an in silico study uncovered the prevalence of a conserved motif (LxxR) in an expanded set of putative pro-domain-containing fungal effectors, some of which can be cleaved by Kex2 in vitro. Our in vitro and in silico study suggests that Kex2-processed pro-domain (designated here as K2PP) effectors are common in fungi and this may have broad implications for the approaches used to study their functions.
Organizational Affiliation:
Research School of Biology, The Australian National University, Canberra, ACT, 2601, Australia.