Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Tan, Y.Z., Zhang, L., Rodrigues, J., Zheng, R.B., Giacometti, S.I., Rosario, A.L., Kloss, B., Dandey, V.P., Wei, H., Brunton, R., Raczkowski, A.M., Athayde, D., Catalao, M.J., Pimentel, M., Clarke, O.B., Lowary, T.L., Archer, M., Niederweis, M., Potter, C.S., Carragher, B., Mancia, F.(2020) Mol Cell 78: 683
- PubMed: 32386575
- DOI: https://doi.org/10.1016/j.molcel.2020.04.014
- Primary Citation of Related Structures:
6W98, 6WBX, 6WBY - PubMed Abstract:
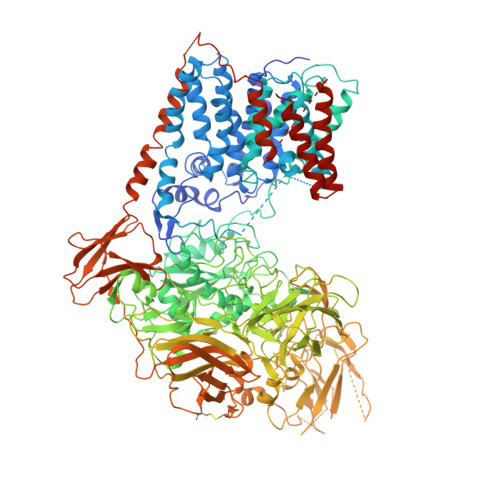
Mycobacterium tuberculosis causes tuberculosis, a disease that kills over 1 million people each year. Its cell envelope is a common antibiotic target and has a unique structure due, in part, to two lipidated polysaccharides-arabinogalactan and lipoarabinomannan. Arabinofuranosyltransferase D (AftD) is an essential enzyme involved in assembling these glycolipids. We present the 2.9-Å resolution structure of M. abscessus AftD, determined by single-particle cryo-electron microscopy. AftD has a conserved GT-C glycosyltransferase fold and three carbohydrate-binding modules. Glycan array analysis shows that AftD binds complex arabinose glycans. Additionally, AftD is non-covalently complexed with an acyl carrier protein (ACP). 3.4- and 3.5-Å structures of a mutant with impaired ACP binding reveal a conformational change, suggesting that ACP may regulate AftD function. Mutagenesis experiments using a conditional knockout constructed in M. smegmatis confirm the essentiality of the putative active site and the ACP binding for AftD function.
Organizational Affiliation:
Department of Physiology and Cellular Biophysics, Columbia University, New York, NY 10032, USA; National Resource for Automated Molecular Microscopy, Simons Electron Microscopy Center, New York Structural Biology Center, New York, NY 10027, USA.