Crystal Structure of Cystathionine beta synthase from Legionella pneumophila with LLP, PLP, and homocysteine
Bolejack, M.J., Davies, D.R., Abendroth, J., Lorimer, D.D., Horanyi, P.S., Edwards, T.E.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cystathionine beta-lyase | 324 | Legionella pneumophila | Mutation(s): 0 Gene Names: C3927_14920, DI056_06925, DI105_06930 | 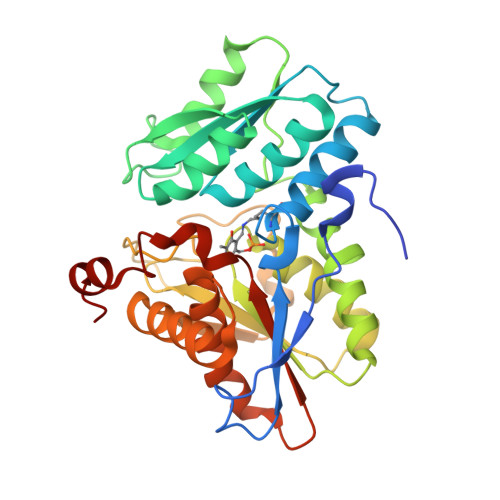 | |
UniProt | |||||
Find proteins for Q5ZRD1 (Legionella pneumophila subsp. pneumophila (strain Philadelphia 1 / ATCC 33152 / DSM 7513)) Explore Q5ZRD1 Go to UniProtKB: Q5ZRD1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5ZRD1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cystathionine beta-lyase | 324 | Legionella pneumophila | Mutation(s): 0 Gene Names: C3927_14920, DI056_06925, DI105_06930 | 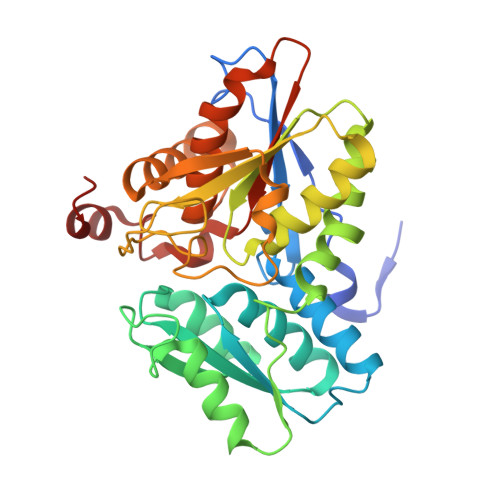 | |
UniProt | |||||
Find proteins for Q5ZRD1 (Legionella pneumophila subsp. pneumophila (strain Philadelphia 1 / ATCC 33152 / DSM 7513)) Explore Q5ZRD1 Go to UniProtKB: Q5ZRD1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5ZRD1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PLP Query on PLP | S [auth B] | PYRIDOXAL-5'-PHOSPHATE C8 H10 N O6 P NGVDGCNFYWLIFO-UHFFFAOYSA-N |  | ||
| HCS Query on HCS | K [auth A] | 2-AMINO-4-MERCAPTO-BUTYRIC ACID C4 H9 N O2 S FFFHZYDWPBMWHY-VKHMYHEASA-N |  | ||
| EDO Query on EDO | C [auth A] D [auth A] E [auth A] H [auth A] M [auth B] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| ACT Query on ACT | F [auth A] G [auth A] I [auth A] J [auth A] L [auth A] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| LLP Query on LLP | A | L-PEPTIDE LINKING | C14 H22 N3 O7 P |  | LYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 65.35 | α = 90 |
| b = 91.79 | β = 90 |
| c = 93.77 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PDB_EXTRACT | data extraction |
| MoRDa | phasing |