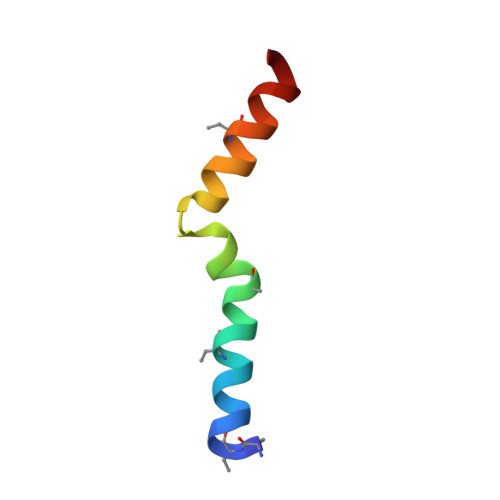
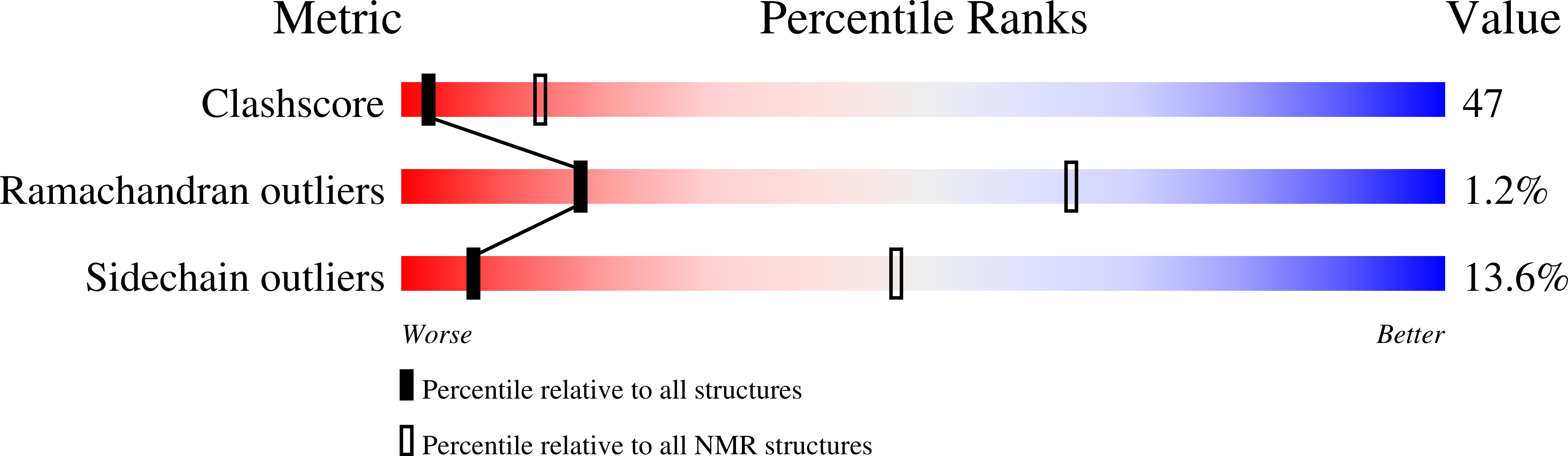Structural determinants of macrocyclization in substrate-controlled lanthipeptide biosynthetic pathways.
Bobeica, S.C., Zhu, L., Acedo, J.Z., Tang, W., van der Donk, W.A.(2020) Chem Sci 11: 12854-12870
- PubMed: 34094481
- DOI: https://doi.org/10.1039/d0sc01651a
- Primary Citation of Related Structures:
6VE9, 6VGT, 6VHJ, 6VJQ, 6VLJ, 7JU9, 7JVF - PubMed Abstract:
Lanthipeptides are characterized by thioether crosslinks formed by post-translational modifications. The cyclization process that favors a single ring pattern over many other possible ring patterns has been the topic of much speculation. Recent studies suggest that for some systems the cyclization pattern and stereochemistry is determined not by the enzyme, but by the sequence of the precursor peptide. However, the factors that govern the outcome of the cyclization process are not understood. This study presents the three-dimensional structures of seven lanthipeptides determined by nuclear magnetic resonance spectroscopy, including five prochlorosins and the two peptides that make up cytolysin, a virulence factor produced by Enterococcus faecalis that is directly linked to human disease. These peptides were chosen because their substrate sequence determines either the ring pattern (prochlorosins) or the stereochemistry of cyclization (cytolysins). We present the structures of prochlorosins 1.1, 2.1, 2.8, 2.10 and 2.11, the first three-dimensional structures of prochlorosins. Our findings provide insights into the molecular determinants of cyclization as well as why some prochlorosins may be better starting points for library generation than others. The structures of the large and small subunits of the enterococcal cytolysin show that these peptides have long helical stretches, a rare observation for lanthipeptides characterized to date. These helices may explain their pore forming activity and suggest that the small subunit may recognize a molecular target followed by recruitment of the large subunit to span the membrane.
Organizational Affiliation:
Department of Chemistry and Howard Hughes Medical Institute, University of Illinois at Urbana-Champaign 600 South Mathews Avenue Urbana Illinois 61801 USA vddonk@illinois.edu +1-217-244-8533 +1-217-244-5360.