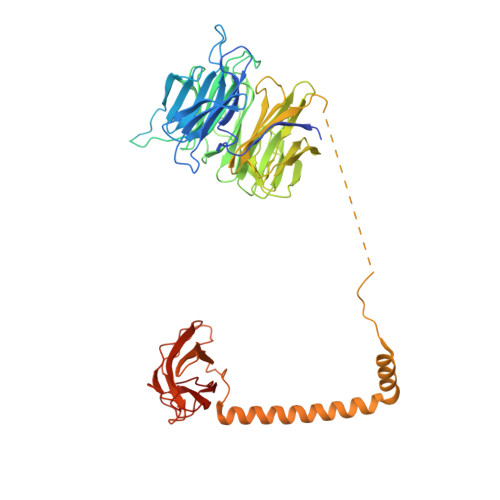
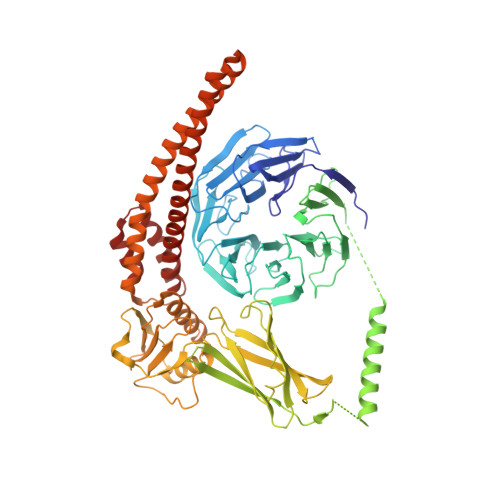
Structure and activation mechanism of the BBSome membrane protein trafficking complex.
Singh, S.K., Gui, M., Koh, F., Yip, M.C., Brown, A.(2020) Elife 9
- PubMed: 31939736
- DOI: https://doi.org/10.7554/eLife.53322
- Primary Citation of Related Structures:
6VBU, 6VBV - PubMed Abstract:
Bardet-Biedl syndrome (BBS) is a currently incurable ciliopathy caused by the failure to correctly establish or maintain cilia-dependent signaling pathways. Eight proteins associated with BBS assemble into the BBSome, a key regulator of the ciliary membrane proteome. We report the electron cryomicroscopy (cryo-EM) structures of the native bovine BBSome in inactive and active states at 3.1 and 3.5 Å resolution, respectively. In the active state, the BBSome is bound to an Arf-family GTPase (ARL6/BBS3) that recruits the BBSome to ciliary membranes. ARL6 recognizes a composite binding site formed by BBS1 and BBS7 that is occluded in the inactive state. Activation requires an unexpected swiveling of the β-propeller domain of BBS1, the subunit most frequently implicated in substrate recognition, which widens a central cavity of the BBSome. Structural mapping of disease-causing mutations suggests that pathogenesis results from folding defects and the disruption of autoinhibition and activation.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Blavatnik Institute, Harvard Medical School, Boston, United States.