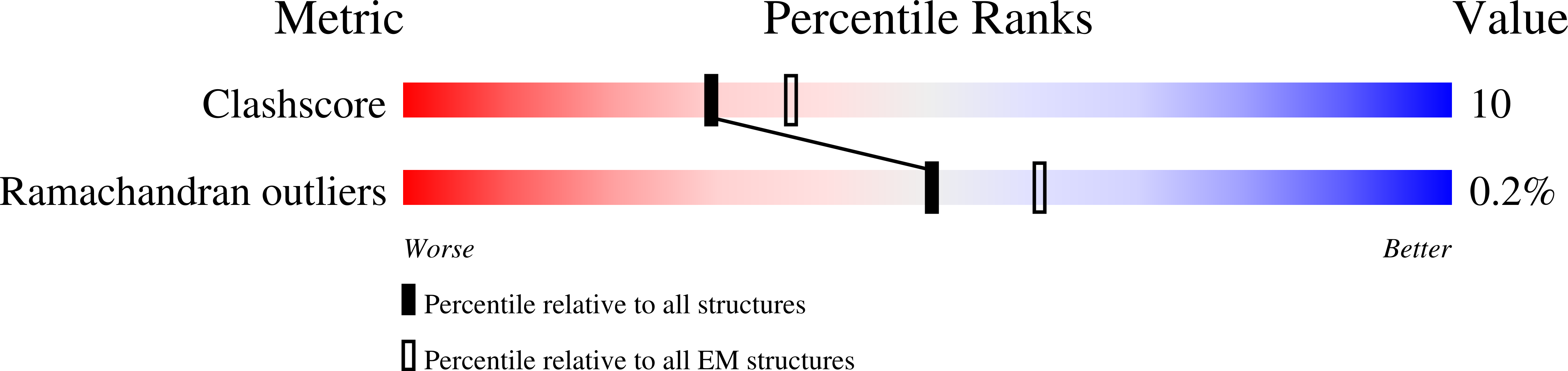Mammalian Retromer Is an Adaptable Scaffold for Cargo Sorting from Endosomes.
Kendall, A.K., Xie, B., Xu, P., Wang, J., Burcham, R., Frazier, M.N., Binshtein, E., Wei, H., Graham, T.R., Nakagawa, T., Jackson, L.P.(2020) Structure 28: 393-405.e4
- PubMed: 32027819
- DOI: https://doi.org/10.1016/j.str.2020.01.009
- Primary Citation of Related Structures:
6VAB, 6VAC - PubMed Abstract:
Metazoan retromer (VPS26/VPS35/VPS29) associates with sorting nexins on endosomal tubules to sort proteins to the trans-Golgi network or plasma membrane. Mechanisms of metazoan retromer assembly remain undefined. We combine single-particle cryoelectron microscopy with biophysical methods to uncover multiple oligomer structures. 2D class averages reveal mammalian heterotrimers; dimers of trimers; tetramers of trimers; and flat chains. These species are further supported by biophysical solution studies. We provide reconstructions of all species, including key sub-structures (∼5 Å resolution). Local resolution variation suggests that heterotrimers and dimers adopt multiple conformations. Our structures identify a flexible, highly conserved electrostatic dimeric interface formed by VPS35 subunits. We generate structure-based mutants to disrupt this interface in vitro. Equivalent mutations in yeast demonstrate a mild cargo-sorting defect. Our data suggest the metazoan retromer is an adaptable and plastic scaffold that accommodates interactions with different sorting nexins to sort multiple cargoes from endosomes their final destinations.
Organizational Affiliation:
Department of Biological Sciences, Vanderbilt University, Nashville, TN 37232, USA; Center for Structural Biology, Vanderbilt University, Nashville, TN 37232, USA.