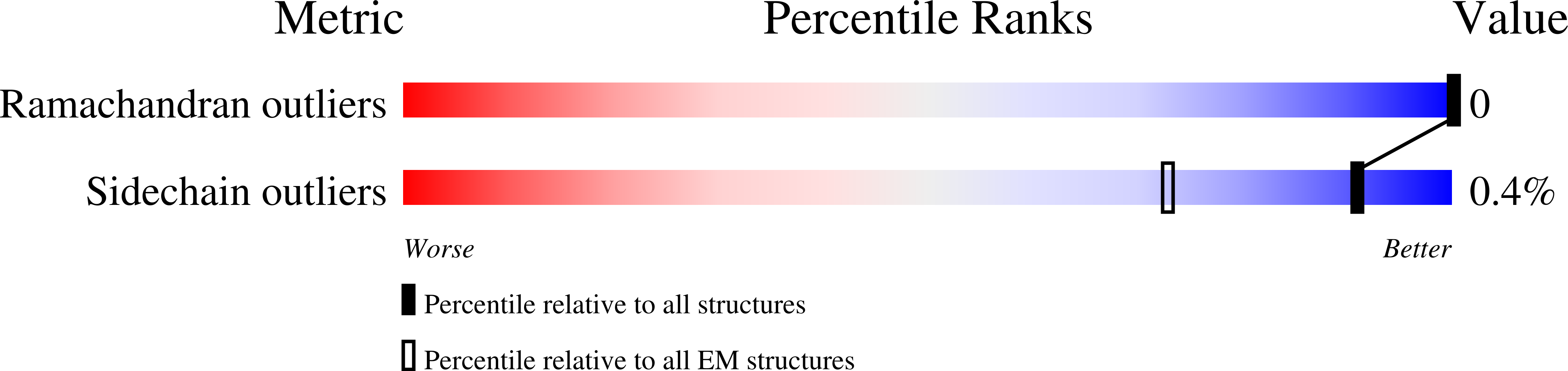Structure of a filamentous virus uncovers familial ties within the archaeal virosphere.
Wang, F., Baquero, D.P., Su, Z., Osinski, T., Prangishvili, D., Egelman, E.H., Krupovic, M.(2020) Virus Evol 6: veaa023-veaa023
- PubMed: 32368353
- DOI: https://doi.org/10.1093/ve/veaa023
- Primary Citation of Related Structures:
6V7B - PubMed Abstract:
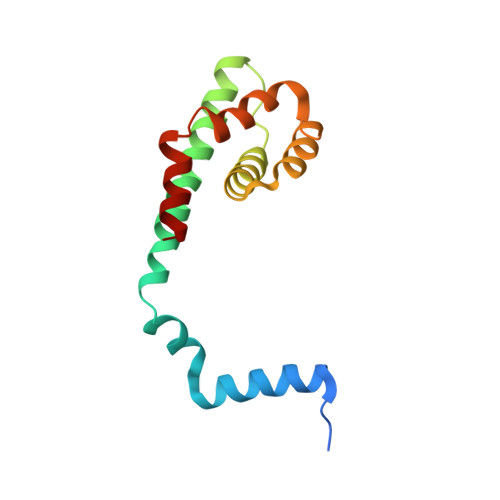
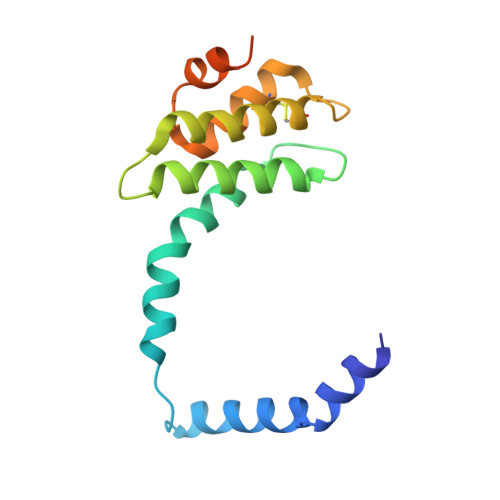
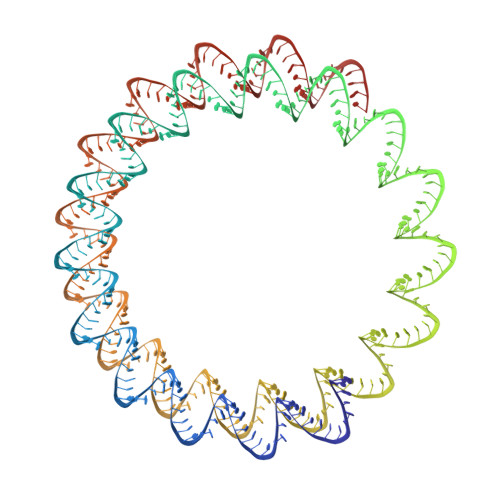
Viruses infecting hyperthermophilic archaea represent one of the most enigmatic parts of the global virome, with viruses from different families showing no genomic relatedness to each other or to viruses of bacteria and eukaryotes. Tristromaviruses, which build enveloped filamentous virions and infect hyperthermophilic neutrophiles of the order Thermoproteales, represent one such enigmatic virus families. They do not share genes with viruses from other families and have been believed to represent an evolutionarily independent virus lineage. A cryo-electron microscopic reconstruction of the tristromavirus Pyrobaculum filamentous virus 2 at 3.4 Å resolution shows that the virion is constructed from two paralogous major capsid proteins (MCP) which transform the linear dsDNA genome of the virus into A-form by tightly wrapping around it. Unexpectedly, the two MCP are homologous to the capsid proteins of other filamentous archaeal viruses, uncovering a deep evolutionary relationship within the archaeal virosphere.
Organizational Affiliation:
Department of Biochemistry and Molecular Genetics, University of Virginia, PO Box 800733, Charlottesville, VA 22908, USA.