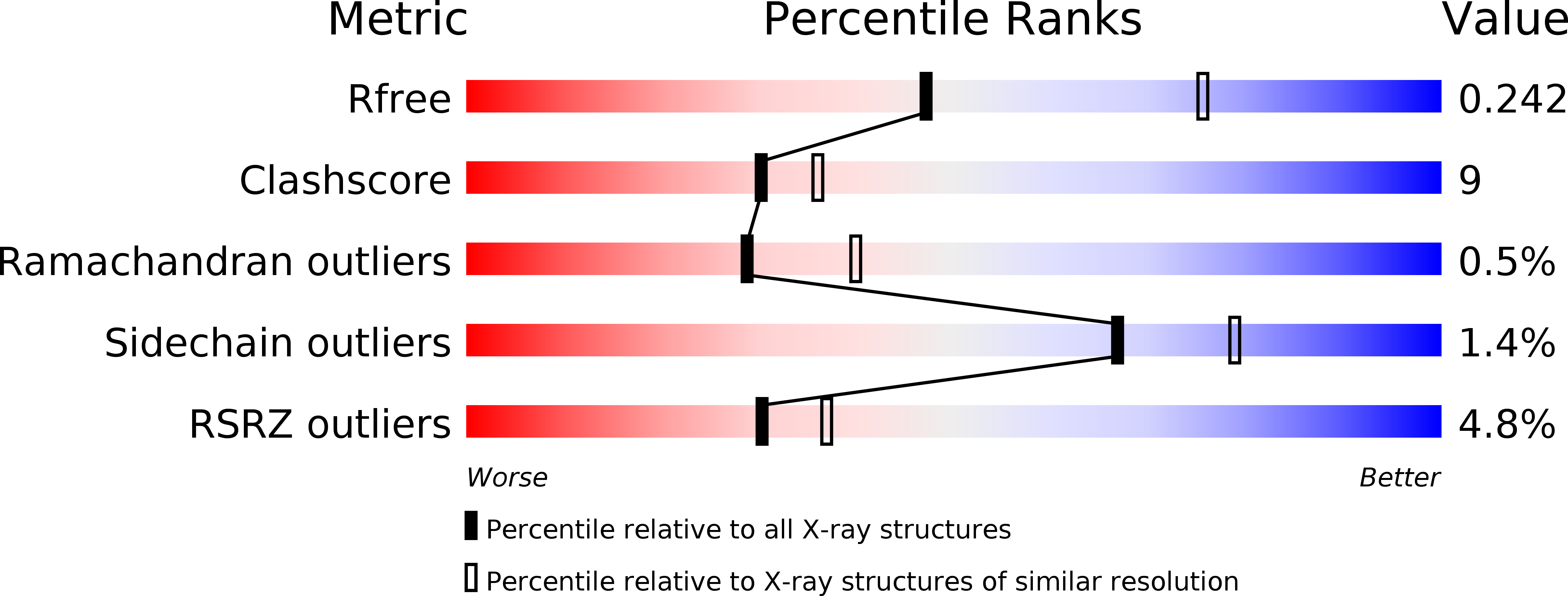
Cytochromecnitrite reductase from the bacteriumGeobacter lovleyirepresents a new NrfA subclass.
Campecino, J., Lagishetty, S., Wawrzak, Z., Sosa Alfaro, V., Lehnert, N., Reguera, G., Hu, J., Hegg, E.L.(2020) J Biol Chem 295: 11455-11465
- PubMed: 32518164
- DOI: https://doi.org/10.1074/jbc.RA120.013981
- Primary Citation of Related Structures:
6V0A - PubMed Abstract:
Cytochrome c nitrite reductase (NrfA) catalyzes the reduction of nitrite to ammonium in the dissimilatory nitrate reduction to ammonium (DNRA) pathway, a process that competes with denitrification, conserves nitrogen, and minimizes nutrient loss in soils. The environmental bacterium Geobacter lovleyi has recently been recognized as a key driver of DNRA in nature, but its enzymatic pathway is still uncharacterized. To address this limitation, here we overexpressed, purified, and characterized G. lovleyi NrfA. We observed that the enzyme crystallizes as a dimer but remains monomeric in solution. Importantly, its crystal structure at 2.55-Å resolution revealed the presence of an arginine residue in the region otherwise occupied by calcium in canonical NrfA enzymes. The presence of EDTA did not affect the activity of G. lovleyi NrfA, and site-directed mutagenesis of this arginine reduced enzymatic activity to <3% of the WT levels. Phylogenetic analysis revealed four separate emergences of Arg-containing NrfA enzymes. Thus, the Ca 2+ -independent, Arg-containing NrfA from G. lovleyi represents a new subclass of cytochrome c nitrite reductase. Most genera from the exclusive clades of Arg-containing NrfA proteins are also represented in clades containing Ca 2+ -dependent enzymes, suggesting convergent evolution.
Organizational Affiliation:
Department of Biochemistry & Molecular Biology, Michigan State University, East Lansing, Michigan, USA.