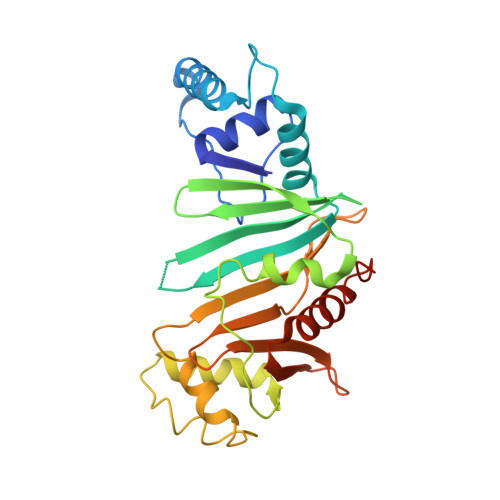
PE5-PPE4-EspG3heterotrimer structure from mycobacterial ESX-3 secretion system gives insight into cognate substrate recognition by ESX systems.
Williamson, Z.A., Chaton, C.T., Ciocca, W.A., Korotkova, N., Korotkov, K.V.(2020) J Biol Chem 295: 12706-12715
- PubMed: 32675282
- DOI: https://doi.org/10.1074/jbc.RA120.012698
- Primary Citation of Related Structures:
6UUJ, 6VHR - PubMed Abstract:
Mycobacterium tuberculosis has evolved numerous type VII secretion (ESX) systems to secrete multiple factors important for both growth and virulence across their cell envelope. ESX-1, ESX-3, and ESX-5 systems have been shown to each secrete a distinct set of substrates, including PE and PPE families of proteins, named for conserved Pro-Glu and Pro-Pro-Glu motifs in their N termini. Proper secretion of the PE-PPE proteins requires the presence of EspG, with each system encoding its own unique copy. There is no cross-talk between any of the ESX systems, and how each EspG recognizes its subset of PE-PPE proteins is currently unknown. The only current structural characterization of PE-PPE-EspG heterotrimers is from the ESX-5 system. Here we present the crystal structure of the PE5 mt -PPE4 mt -EspG 3mm heterotrimer from the ESX-3 system. Our heterotrimer reveals that EspG 3mm interacts exclusively with PPE4 mt in a similar manner to EspG 5 , shielding the hydrophobic tip of PPE4 mt from solvent. The C-terminal helical domain of EspG 3mm is dynamic, alternating between "open" and "closed" forms, and this movement is likely functionally relevant in the unloading of PE-PPE heterodimers at the secretion machinery. In contrast to the previously solved ESX-5 heterotrimers, the PE-PPE heterodimer of our ESX-3 heterotrimer is interacting with its chaperone at a drastically different angle and presents different faces of the PPE protein to the chaperone. We conclude that the PPE-EspG interface from each ESX system has a unique shape complementarity that allows each EspG to discriminate among noncognate PE-PPE pairs.
Organizational Affiliation:
Department of Molecular & Cellular Biochemistry and the Center for Structural Biology, University of Kentucky, Lexington, Kentucky, USA.