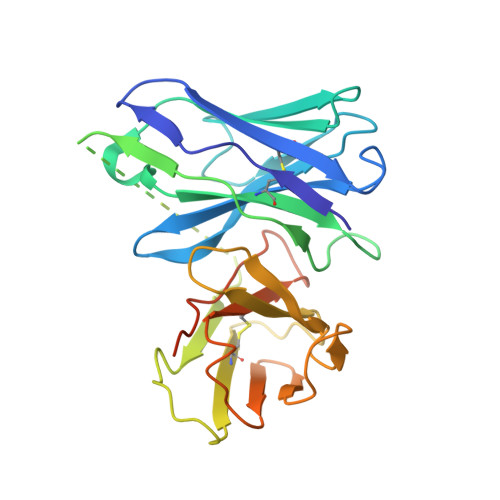
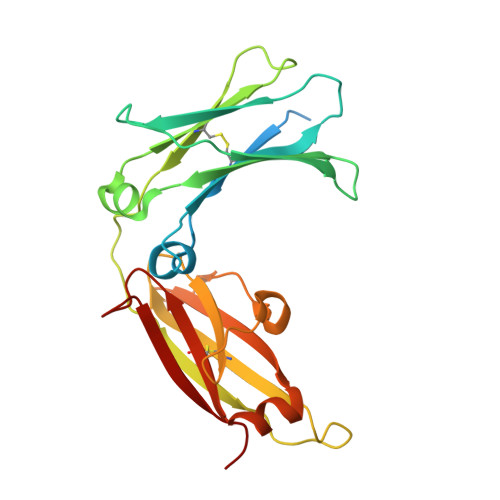
The mechanistic and functional profile of the therapeutic anti-IgE antibody ligelizumab differs from omalizumab.
Gasser, P., Tarchevskaya, S.S., Guntern, P., Brigger, D., Ruppli, R., Zbaren, N., Kleinboelting, S., Heusser, C., Jardetzky, T.S., Eggel, A.(2020) Nat Commun 11: 165-165
- PubMed: 31913280
- DOI: https://doi.org/10.1038/s41467-019-13815-w
- Primary Citation of Related Structures:
6UQR - PubMed Abstract:
Targeting of immunoglobulin E (IgE) represents an interesting approach for the treatment of allergic disorders. A high-affinity monoclonal anti-IgE antibody, ligelizumab, has recently been developed to overcome some of the limitations associated with the clinical use of the therapeutic anti-IgE antibody, omalizumab. Here, we determine the molecular binding profile and functional modes-of-action of ligelizumab. We solve the crystal structure of ligelizumab bound to IgE, and report epitope differences between ligelizumab and omalizumab that contribute to their qualitatively distinct IgE-receptor inhibition profiles. While ligelizumab shows superior inhibition of IgE binding to FcεRI, basophil activation, IgE production by B cells and passive systemic anaphylaxis in an in vivo mouse model, ligelizumab is less potent in inhibiting IgE:CD23 interactions than omalizumab. Our data thus provide a structural and mechanistic foundation for understanding the efficient suppression of FcεRI-dependent allergic reactions by ligelizumab in vitro as well as in vivo.
Organizational Affiliation:
Department of BioMedical Research, University of Bern, Bern, Switzerland.