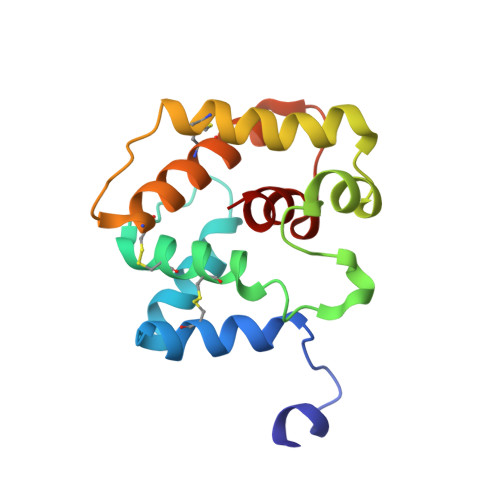
Ligand- and pH-Induced Structural Transition of Gypsy Moth Lymantria dispar Pheromone-Binding Protein 1 (LdisPBP1).
Terrado, M., Okon, M., McIntosh, L.P., Plettner, E.(2020) Biochemistry 59: 3411-3426
- PubMed: 32877603
- DOI: https://doi.org/10.1021/acs.biochem.0c00592
- Primary Citation of Related Structures:
6UM9 - PubMed Abstract:
Pheromone-binding proteins (PBPs) are small, water-soluble proteins found in the lymph of pheromone-sensing hairs. PBPs are essential in modulating pheromone partitioning in the lymph and at pheromone receptors of olfactory sensory neurons. The function of a PBP is associated with its ability to structurally convert between two conformations. Although mechanistic details remain unclear, it has been proposed that the structural transition between these forms is affected by two factors: pH and the presence or absence of ligand. To better understand the PBP conformational transition, the structure of the gypsy moth ( Lymantria dispar ) LdisPBP1 was elucidated at pH 4.5 and 35 °C using nuclear magnetic resonance spectroscopy. In addition, the effects of sample pH and binding of the species' pheromone, (+)-disparlure, (7 R ,8 S )-epoxy-2-methyloctadecane, and its enantiomer were monitored via 15 N HSQC spectroscopy. LdisPBP1 in acidic conditions has seven helices, with its C-terminal residues forming the seventh helix within the pheromone-binding pocket and its N-terminal residues disordered. Under conditions where this conformation is made favorable, free LdisPBP1 would have limited ligand binding capacity due to the seventh helix occupying the internal binding pocket. Our findings suggest that even in the presence of 4-fold ligand at acidic pH, LdisPBP1 is only ∼60% in its pheromone-bound form. Furthermore, evidence of a different LdisPBP1 form is seen at higher pH, with the transition pH between 5.6 and 6.0. This suggests that LdisPBP1 at neutral pH exists as a mixture of at least two conformations. These findings have implications concerning the PBP ligand binding and release mechanism.
Organizational Affiliation:
Department of Chemistry, Simon Fraser University, Burnaby, British Columbia V5A 1S6, Canada.