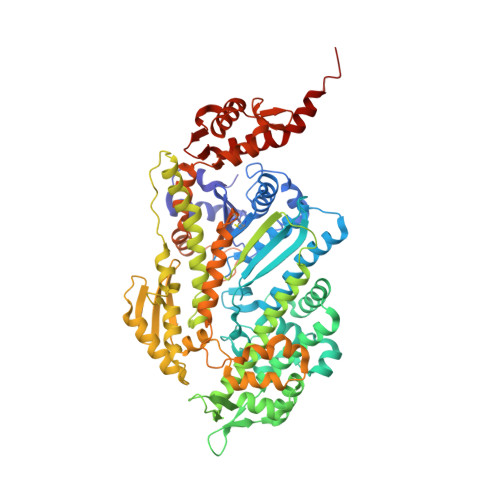
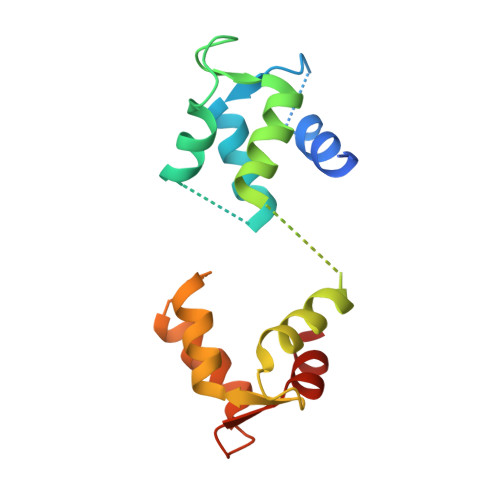
Structural basis of Fusarium myosin I inhibition by phenamacril.
Zhou, Y., Zhou, X.E., Gong, Y., Zhu, Y., Cao, X., Brunzelle, J.S., Xu, H.E., Zhou, M., Melcher, K., Zhang, F.(2020) PLoS Pathog 16: e1008323-e1008323
- PubMed: 32163521
- DOI: https://doi.org/10.1371/journal.ppat.1008323
- Primary Citation of Related Structures:
6UI4 - PubMed Abstract:
Fusarium is a genus of filamentous fungi that includes species that cause devastating diseases in major staple crops, such as wheat, maize, rice, and barley, resulting in severe yield losses and mycotoxin contamination of infected grains. Phenamacril is a novel fungicide that is considered environmentally benign due to its exceptional specificity; it inhibits the ATPase activity of the sole class I myosin of only a subset of Fusarium species including the major plant pathogens F. graminearum, F. asiaticum and F. fujikuroi. To understand the underlying mechanisms of inhibition, species specificity, and resistance mutations, we have determined the crystal structure of phenamacril-bound F. graminearum myosin I. Phenamacril binds in the actin-binding cleft in a new allosteric pocket that contains the central residue of the regulatory Switch 2 loop and that is collapsed in the structure of a myosin with closed actin-binding cleft, suggesting that pocket occupancy blocks cleft closure. We have further identified a single, transferable phenamacril-binding residue found exclusively in phenamacril-sensitive myosins to confer phenamacril selectivity.
Organizational Affiliation:
Key Laboratory of Pesticide, College of Plant Protection, Nanjing Agricultural University, Nanjing, China.