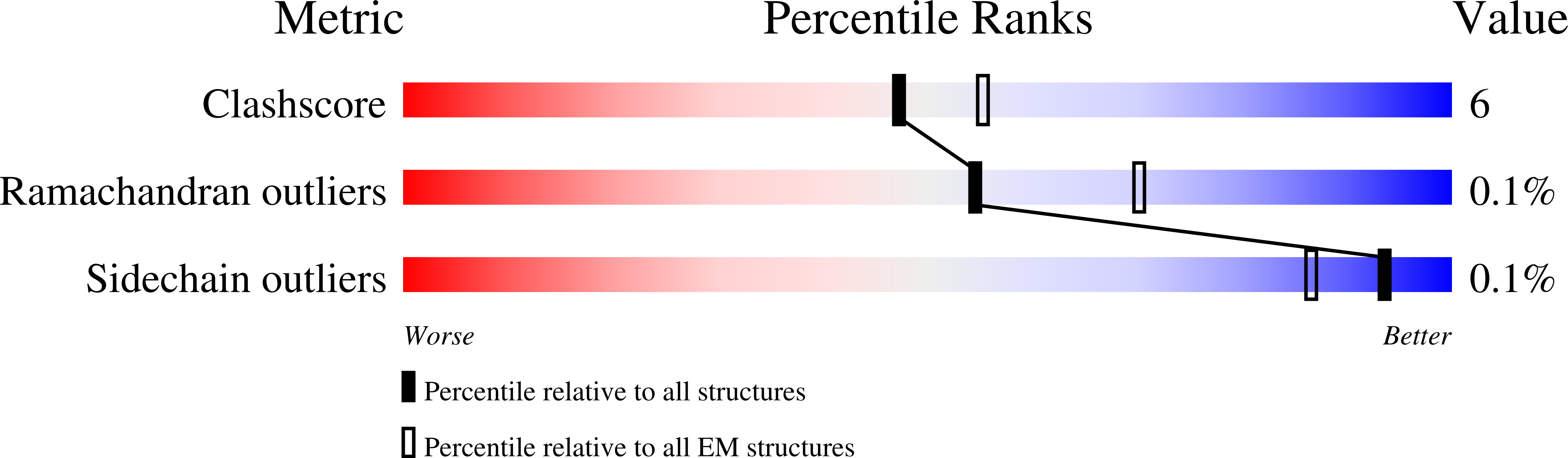Structure of a rabies virus polymerase complex from electron cryo-microscopy.
Horwitz, J.A., Jenni, S., Harrison, S.C., Whelan, S.P.J.(2020) Proc Natl Acad Sci U S A 117: 2099-2107
- PubMed: 31953264
- DOI: https://doi.org/10.1073/pnas.1918809117
- Primary Citation of Related Structures:
6UEB - PubMed Abstract:
Nonsegmented negative-stranded (NNS) RNA viruses, among them the virus that causes rabies (RABV), include many deadly human pathogens. The large polymerase (L) proteins of NNS RNA viruses carry all of the enzymatic functions required for viral messenger RNA (mRNA) transcription and replication: RNA polymerization, mRNA capping, and cap methylation. We describe here a complete structure of RABV L bound with its phosphoprotein cofactor (P), determined by electron cryo-microscopy at 3.3 Å resolution. The complex closely resembles the vesicular stomatitis virus (VSV) L-P, the one other known full-length NNS-RNA L-protein structure, with key local differences (e.g., in L-P interactions). Like the VSV L-P structure, the RABV complex analyzed here represents a preinitiation conformation. Comparison with the likely elongation state, seen in two structures of pneumovirus L-P complexes, suggests differences between priming/initiation and elongation complexes. Analysis of internal cavities within RABV L suggests distinct template and product entry and exit pathways during transcription and replication.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115.