Fungal-derived brevianamide assembly by a stereoselective semipinacolase.
Ye, Y., Du, L., Zhang, X., Newmister, S.A., McCauley, M., Alegre-Requena, J.V., Zhang, W., Mu, S., Minami, A., Fraley, A.E., Adrover-Castellano, M.L., Carney, N.A., Shende, V.V., Qi, F., Oikawa, H., Kato, H., Tsukamoto, S., Paton, R.S., Williams, R.M., Sherman, D.H., Li, S.(2020) Nat Catal 3: 497-506
- PubMed: 32923978
- DOI: https://doi.org/10.1038/s41929-020-0454-9
- Primary Citation of Related Structures:
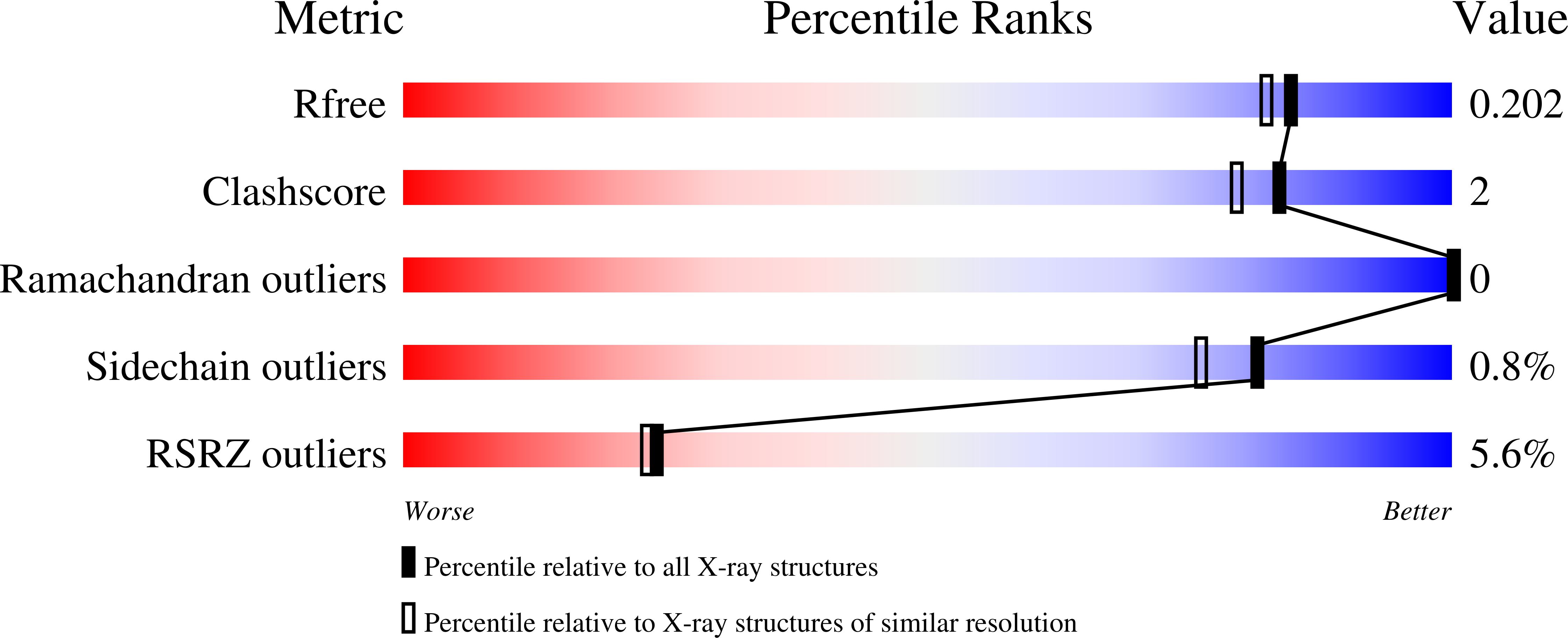
6U9I - PubMed Abstract:
Fungal bicyclo[2.2.2]diazaoctane indole alkaloids represent an important family of natural products with a wide-spectrum of biological activities. Although biomimetic total syntheses of representative compounds have been reported, the details of their biogenesis, especially the mechanisms for assembly of diastereomerically distinct and enantiomerically antipodal metabolites, have remained largely uncharacterized. Brevianamide A represents a basic form of the sub-family bearing a dioxopiperazine core and a rare 3- spiro -ψ-indoxyl skeleton. Here, we identified the Brevianamide A biosynthetic gene cluster from Penicillium brevicompactum NRRL 864 and elucidated the metabolic pathway. BvnE was revealed to be an essential isomerase/semi-pinacolase that specifies selective production of the natural product. Structural elucidation, molecular modeling, and mutational analysis of BvnE, and quantum chemical calculations provided mechanistic insights into the diastereoselective formation of the 3- spiro -ψ-indoxyl moiety in Brevianamide A. This occurs through a BvnE-controlled semi-pinacol rearrangement and a subsequent spontaneous intramolecular [4+2] hetero -Diels-Alder cycloaddition.
Organizational Affiliation:
Life Sciences Institute, University of Michigan, Ann Arbor, Michigan 48109, USA.