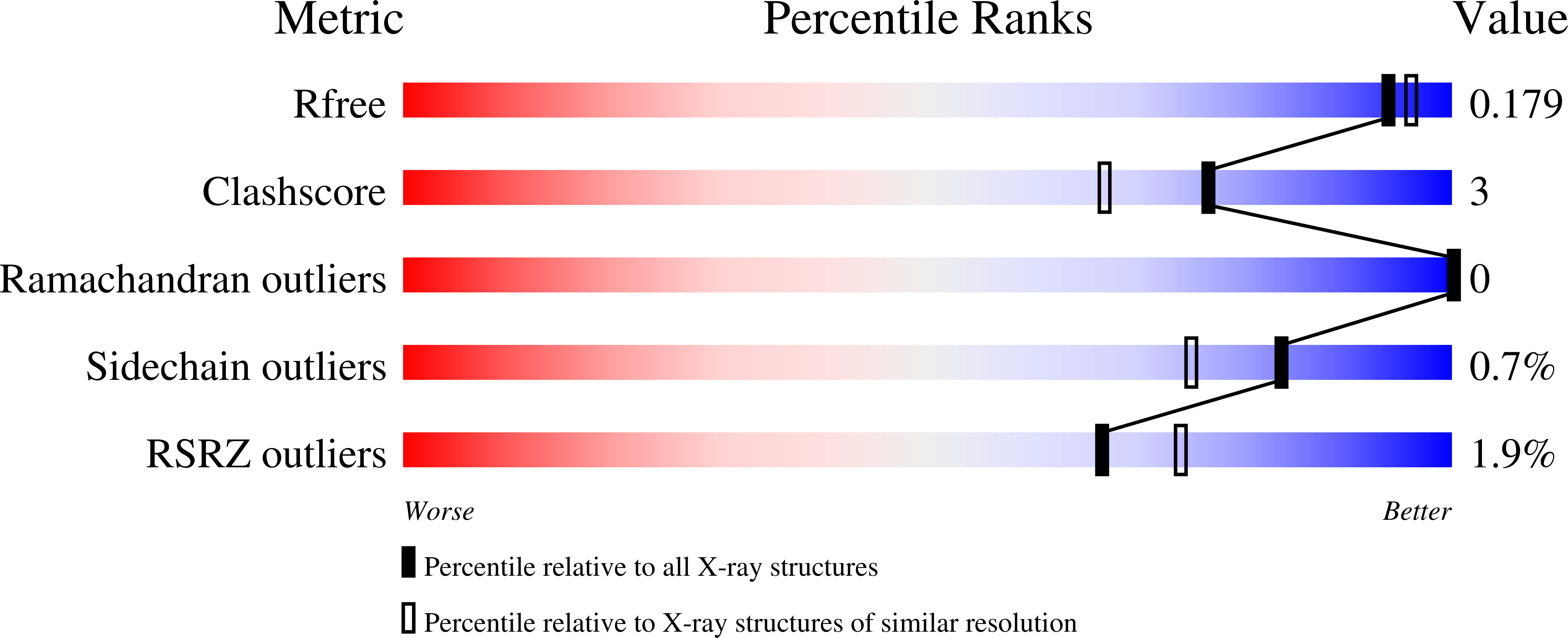
Structural Mechanism of Regioselectivity in an Unusual Bacterial Acyl-CoA Dehydrogenase.
Blake-Hedges, J.M., Pereira, J.H., Cruz-Morales, P., Thompson, M.G., Barajas, J.F., Chen, J., Krishna, R.N., Chan, L.J.G., Nimlos, D., Alonso-Martinez, C., Baidoo, E.E.K., Chen, Y., Gin, J.W., Katz, L., Petzold, C.J., Adams, P.D., Keasling, J.D.(2020) J Am Chem Soc 142: 835-846
- PubMed: 31793780
- DOI: https://doi.org/10.1021/jacs.9b09187
- Primary Citation of Related Structures:
6U1V - PubMed Abstract:
Terminal alkenes are easily derivatized, making them desirable functional group targets for polyketide synthase (PKS) engineering. However, they are rarely encountered in natural PKS systems. One mechanism for terminal alkene formation in PKSs is through the activity of an acyl-CoA dehydrogenase (ACAD). Herein, we use biochemical and structural analysis to understand the mechanism of terminal alkene formation catalyzed by an γ,δ-ACAD from the biosynthesis of the polyketide natural product FK506, TcsD. While TcsD is homologous to canonical α,β-ACADs, it acts regioselectively at the γ,δ-position and only on α,β-unsaturated substrates. Furthermore, this regioselectivity is controlled by a combination of bulky residues in the active site and a lateral shift in the positioning of the FAD cofactor within the enzyme. Substrate modeling suggests that TcsD utilizes a novel set of hydrogen bond donors for substrate activation and positioning, preventing dehydrogenation at the α,β position of substrates. From the structural and biochemical characterization of TcsD, key residues that contribute to regioselectivity and are unique to the protein family were determined and used to identify other putative γ,δ-ACADs that belong to diverse natural product biosynthetic gene clusters. These predictions are supported by the demonstration that a phylogenetically distant homologue of TcsD also regioselectively oxidizes α,β-unsaturated substrates. This work exemplifies a powerful approach to understand unique enzymatic reactions and will facilitate future enzyme discovery, inform enzyme engineering, and aid natural product characterization efforts.
Organizational Affiliation:
Department of Chemistry , University of California , Berkeley , California 94720 , United States.