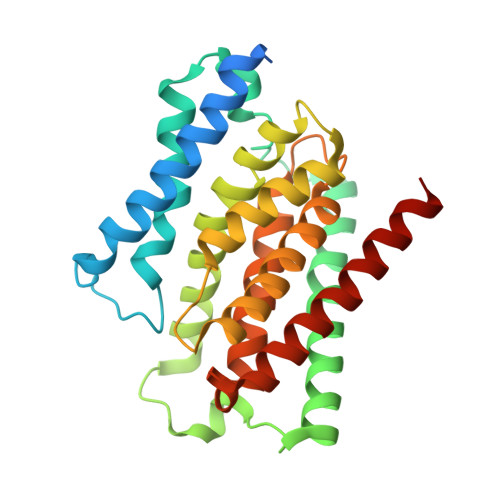
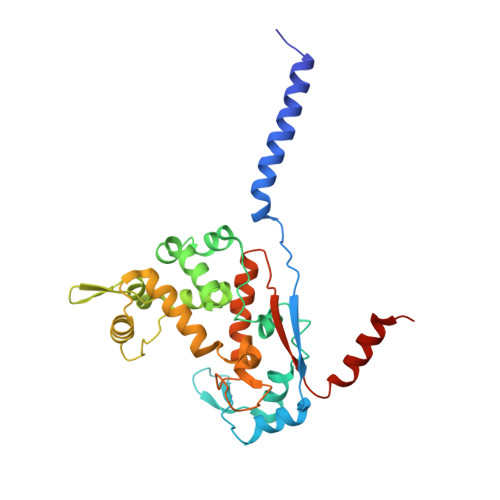
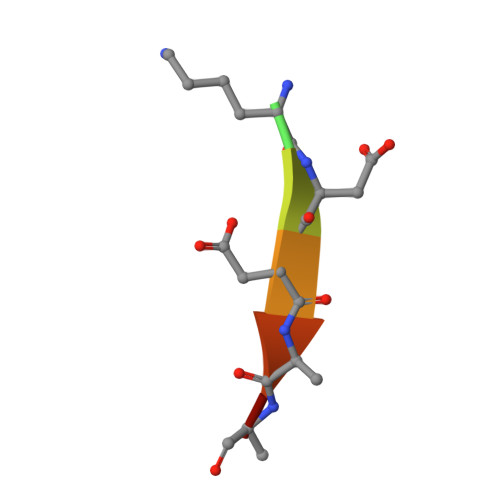
Structure and reconstitution of a hydrolase complex that may release peptidoglycan from the membrane after polymerization.
Schaefer, K., Owens, T.W., Page, J.E., Santiago, M., Kahne, D., Walker, S.(2021) Nat Microbiol 6: 34-43
- PubMed: 33168989
- DOI: https://doi.org/10.1038/s41564-020-00808-5
- Primary Citation of Related Structures:
6U0O - PubMed Abstract:
Bacteria are encapsulated by a peptidoglycan cell wall that is essential for their survival 1 . During cell wall assembly, a lipid-linked disaccharide-peptide precursor called lipid II is polymerized and cross-linked to produce mature peptidoglycan. As lipid II is polymerized, nascent polymers remain membrane-anchored at one end, and the other end becomes cross-linked to the matrix 2-4 . How bacteria release newly synthesized peptidoglycan strands from the membrane to complete the synthesis of mature peptidoglycan is a long-standing question. Here, we show that a Staphylococcus aureus cell wall hydrolase and a membrane protein that contains eight transmembrane helices form a complex that may function as a peptidoglycan release factor. The complex cleaves nascent peptidoglycan internally to produce free oligomers as well as lipid-linked oligomers that can undergo further elongation. The polytopic membrane protein, which is similar to a eukaryotic CAAX protease, controls the length of these products. A structure of the complex at a resolution of 2.6 Å shows that the membrane protein scaffolds the hydrolase to orient its active site for cleaving the glycan strand. We propose that this complex functions to detach newly synthesized peptidoglycan polymer from the cell membrane to complete integration into the cell wall matrix.
Organizational Affiliation:
Department of Microbiology, Harvard Medical School, Boston, MA, USA.