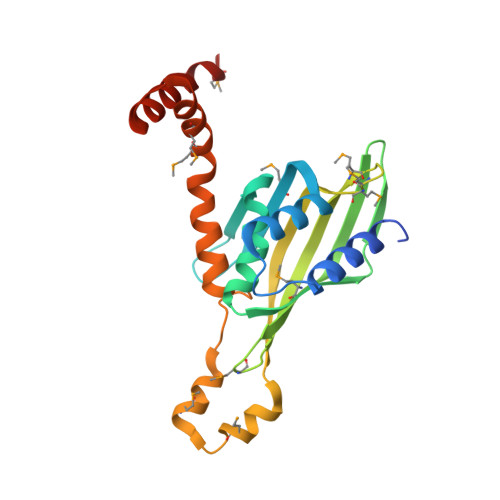
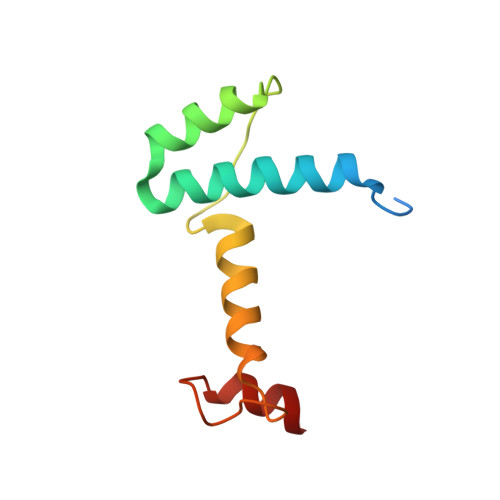
Crystal structure of human LDB1 in complex with SSBP2.
Wang, H., Kim, J., Wang, Z., Yan, X.X., Dean, A., Xu, W.(2020) Proc Natl Acad Sci U S A 117: 1042-1048
- PubMed: 31892537
- DOI: https://doi.org/10.1073/pnas.1914181117
- Primary Citation of Related Structures:
6TYD - PubMed Abstract:
The Lim domain binding proteins (LDB1 and LDB2 in human and Chip in Drosophila ) play critical roles in cell fate decisions through partnership with multiple Lim-homeobox and Lim-only proteins in diverse developmental systems including cardiogenesis, neurogenesis, and hematopoiesis. In mammalian erythroid cells, LDB1 dimerization supports long-range connections between enhancers and genes involved in erythropoiesis, including the β-globin genes. Single-stranded DNA binding proteins (SSBPs) interact specifically with the LDB/Chip conserved domain (LCCD) of LDB proteins and stabilize LDBs by preventing their proteasomal degradation, thus promoting their functions in gene regulation. The structural basis for LDB1 self-interaction and interface with SSBPs is unclear. Here we report a crystal structure of the human LDB1/SSBP2 complex at 2.8-Å resolution. The LDB1 dimerization domain (DD) contains an N-terminal nuclear transport factor 2 (NTF2)-like subdomain and a small helix 4-helix 5 subdomain, which together form the LDB1 dimerization interface. The 2 LCCDs in the symmetric LDB1 dimer flank the core DDs, with each LCCD forming extensive interactions with an SSBP2 dimer. The conserved linker between LDB1 DD and LCCD covers a potential ligand-binding pocket of the LDB1 NTF2-like subdomain and may serve as a regulatory site for LDB1 structure and function. Our structural and biochemical data provide a much-anticipated structural basis for understanding how LDB1 and the LDB1/SSBP interactions form the structural core of diverse complexes mediating cell choice decisions and long-range enhancer-promoter interactions.
Organizational Affiliation:
National Laboratory of Biomacromolecules, Chinese Academy of Sciences Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, 100101 Beijing, China.