How to limit the speed of a motor: the intricate regulation of the XPB ATPase and translocase in TFIIH.
Kappenberger, J., Koelmel, W., Schoenwetter, E., Scheuer, T., Woerner, J., Kuper, J., Kisker, C.(2020) Nucleic Acids Res 48: 12282-12296
- PubMed: 33196848
- DOI: https://doi.org/10.1093/nar/gkaa911
- Primary Citation of Related Structures:
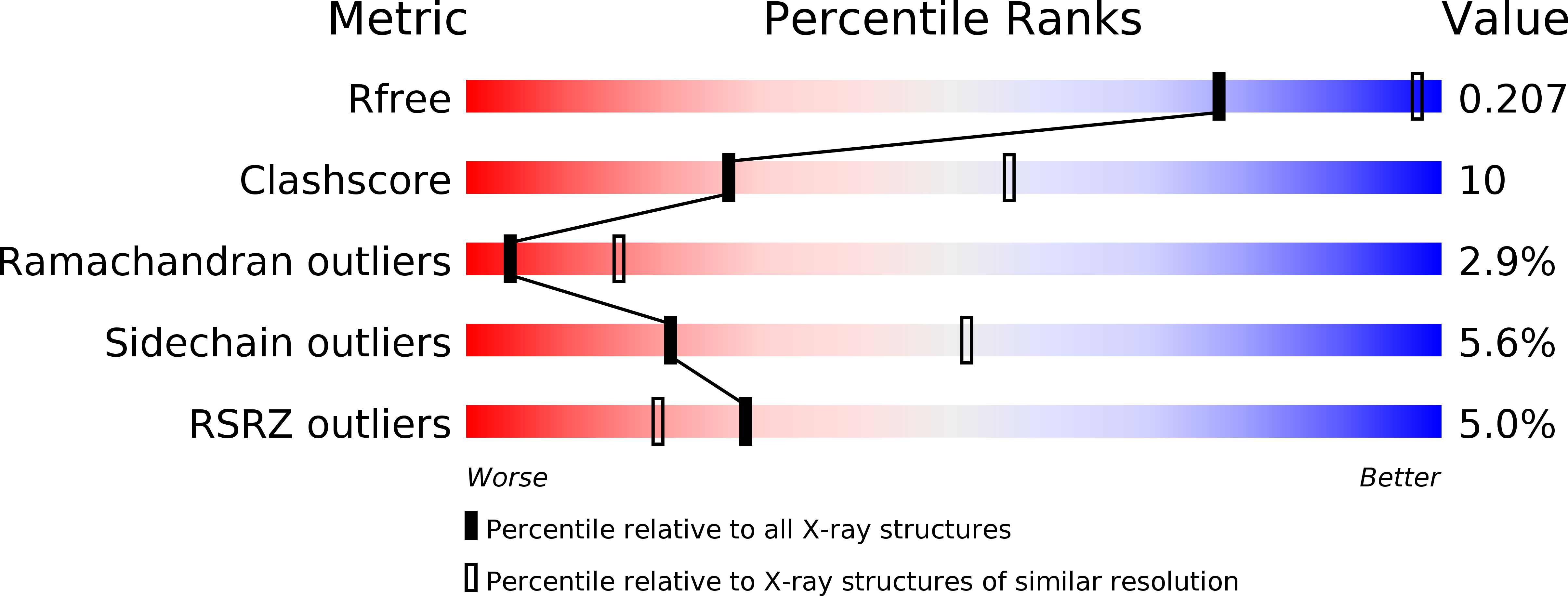
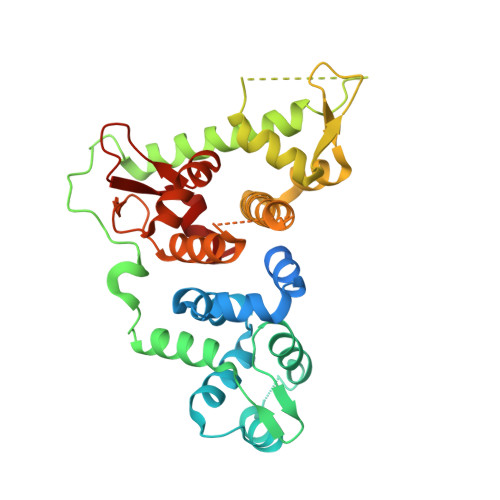
6TRS, 6TRU - PubMed Abstract:
The superfamily 2 helicase XPB is an integral part of the general transcription factor TFIIH and assumes essential catalytic functions in transcription initiation and nucleotide excision repair. The ATPase activity of XPB is required in both processes. We investigated the interaction network that regulates XPB via the p52 and p8 subunits with functional mutagenesis based on our crystal structure of the p52/p8 complex and current cryo-EM structures. Importantly, we show that XPB's ATPase can be activated either by DNA or by the interaction with the p52/p8 proteins. Intriguingly, we observe that the ATPase activation by p52/p8 is significantly weaker than the activation by DNA and when both p52/p8 and DNA are present, p52/p8 dominates the maximum activation. We therefore define p52/p8 as the master regulator of XPB acting as an activator and speed limiter at the same time. A correlative analysis of the ATPase and translocase activities of XPB shows that XPB only acts as a translocase within the context of complete core TFIIH and that XPA increases the processivity of the translocase complex without altering XPB's ATPase activity. Our data define an intricate network that tightly controls the activity of XPB during transcription and nucleotide excision repair.
Organizational Affiliation:
Rudolf Virchow Center for Integrative and Translational Bioimaging, Institute for Structural Biology, University of Würzburg, 97080 Würzburg, Germany.