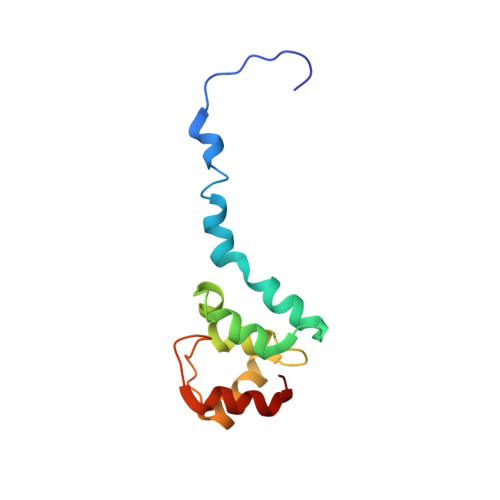
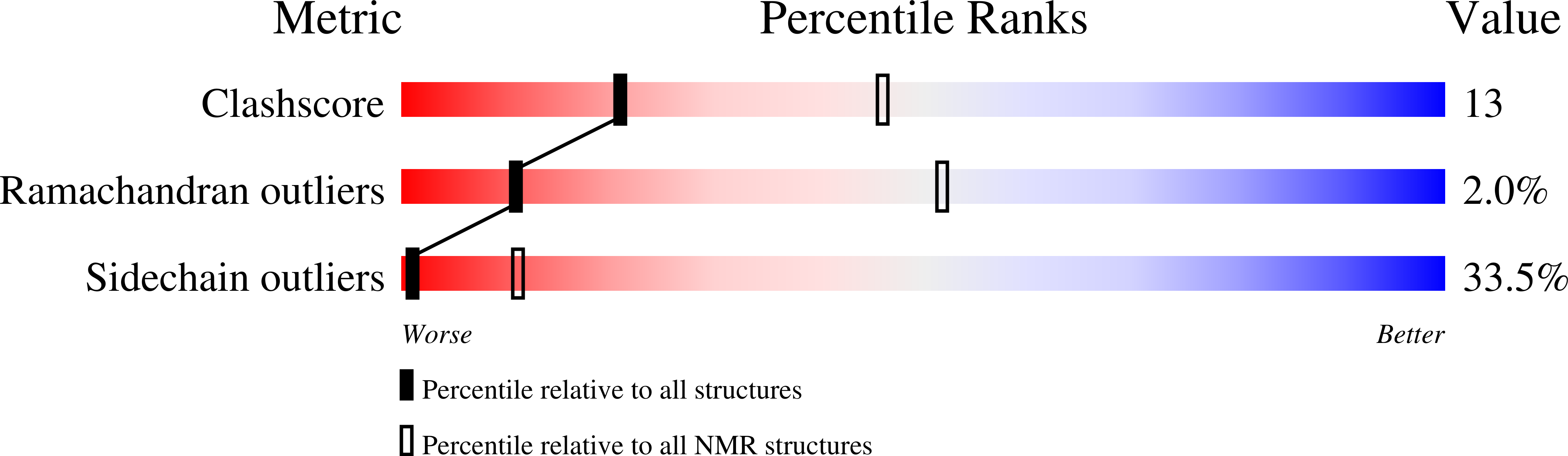Structural and DNA binding properties of mycobacterial integration host factor mIHF.
Odermatt, N.T., Lelli, M., Herrmann, T., Abriata, L.A., Japaridze, A., Voilquin, H., Singh, R., Piton, J., Emsley, L., Dietler, G., Cole, S.T.(2020) J Struct Biol 209: 107434-107434
- PubMed: 31846718
- DOI: https://doi.org/10.1016/j.jsb.2019.107434
- Primary Citation of Related Structures:
6TOB - PubMed Abstract:
In bacteria, nucleoid associated proteins (NAPs) take part in active chromosome organization by supercoil management, three-dimensional DNA looping and direct transcriptional control. Mycobacterial integration host factor (mIHF, rv1388) is a NAP restricted to Actinobacteria and essential for survival of the human pathogen Mycobacterium tuberculosis. We show in vitro that DNA binding by mIHF strongly stabilizes the protein and increases its melting temperature. The structure obtained by Nuclear Magnetic Resonance (NMR) spectroscopy characterizes mIHF as a globular protein with a protruding alpha helix and a disordered N-terminus, similar to Streptomyces coelicolor IHF (sIHF). NMR revealed no residues of high flexibility, suggesting that mIHF is a rigid protein overall that does not undergo structural rearrangements. We show that mIHF only binds to double stranded DNA in solution, through two DNA binding sites (DBSs) similar to those identified in the X-ray structure of sIHF. According to Atomic Force Microscopy, mIHF is able to introduce left-handed loops of ca. 100 nm size (~300 bp) in supercoiled cosmids, thereby unwinding and relaxing the DNA.
Organizational Affiliation:
École Polytechnique Fédérale de Lausanne, School of Life Sciences, Station 19, 1015 Lausanne, Switzerland. Electronic address: nina.odermatt@mpi-marburg.mpg.de.