Triple-Helix-Stabilizing Effects in Collagen Model Peptides Containing PPII-Helix-Preorganized Diproline Modules.
Maassen, A., Gebauer, J.M., Theres Abraham, E., Grimm, I., Neudorfl, J.M., Kuhne, R., Neundorf, I., Baumann, U., Schmalz, H.G.(2020) Angew Chem Int Ed Engl 59: 5747-5755
- PubMed: 31944532
- DOI: https://doi.org/10.1002/anie.201914101
- Primary Citation of Related Structures:
6SYJ - PubMed Abstract:
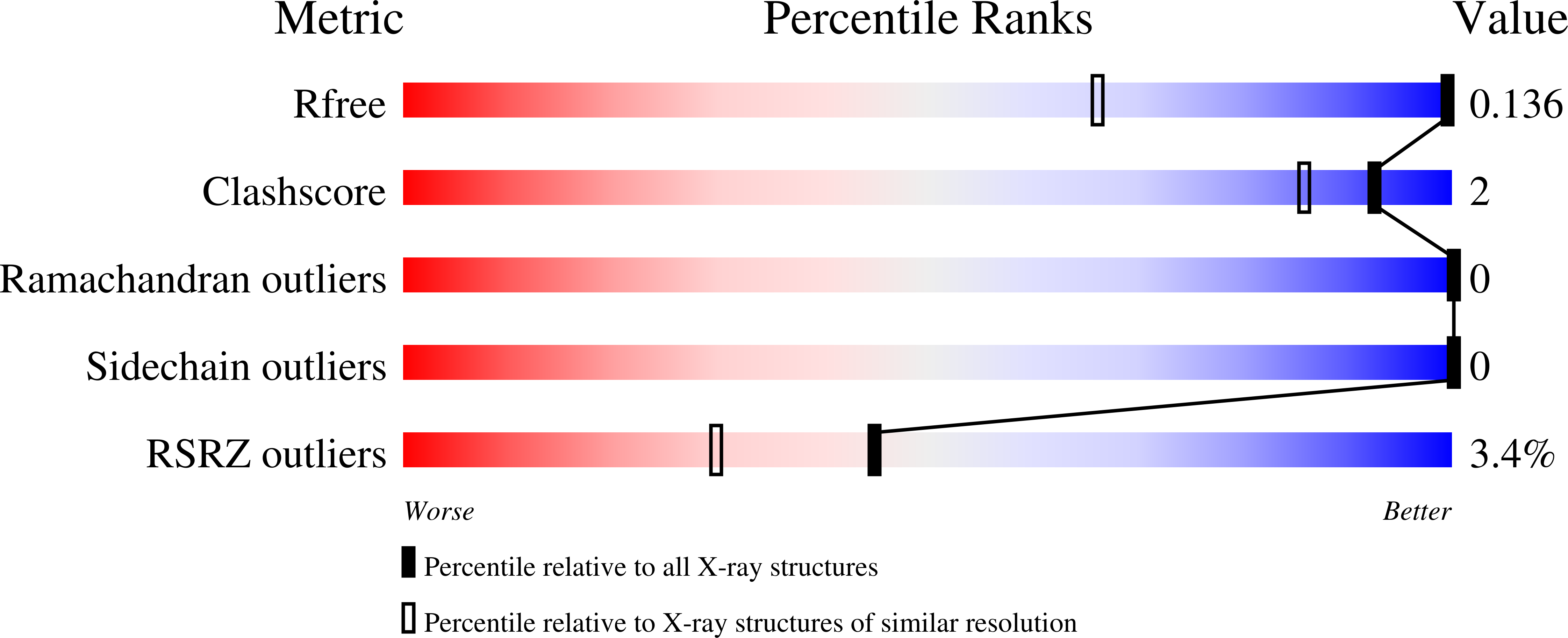
Collagen model peptides (CMPs) serve as tools for understanding stability and function of the collagen triple helix and have a potential for biomedical applications. In the past, interstrand cross-linking or conformational preconditioning of proline units through stereoelectronic effects have been utilized in the design of stabilized CMPs. To further study the effects determining collagen triple helix stability we investigated a series of CMPs containing synthetic diproline-mimicking modules (ProMs), which were preorganized in a PPII-helix-type conformation by a functionalizable intrastrand C 2 bridge. Results of CD-based denaturation studies were correlated with calculated (DFT) conformational preferences of the ProM units, revealing that the relative helix stability is mainly governed by an interplay of main-chain preorganization, ring-flip preference, adaptability, and steric effects. Triple helix integrity was proven by crystal structure analysis and binding to HSP47.
Organizational Affiliation:
University of Cologne, Department of Chemistry, Greinstraße 4, 50939, Cologne, Germany.