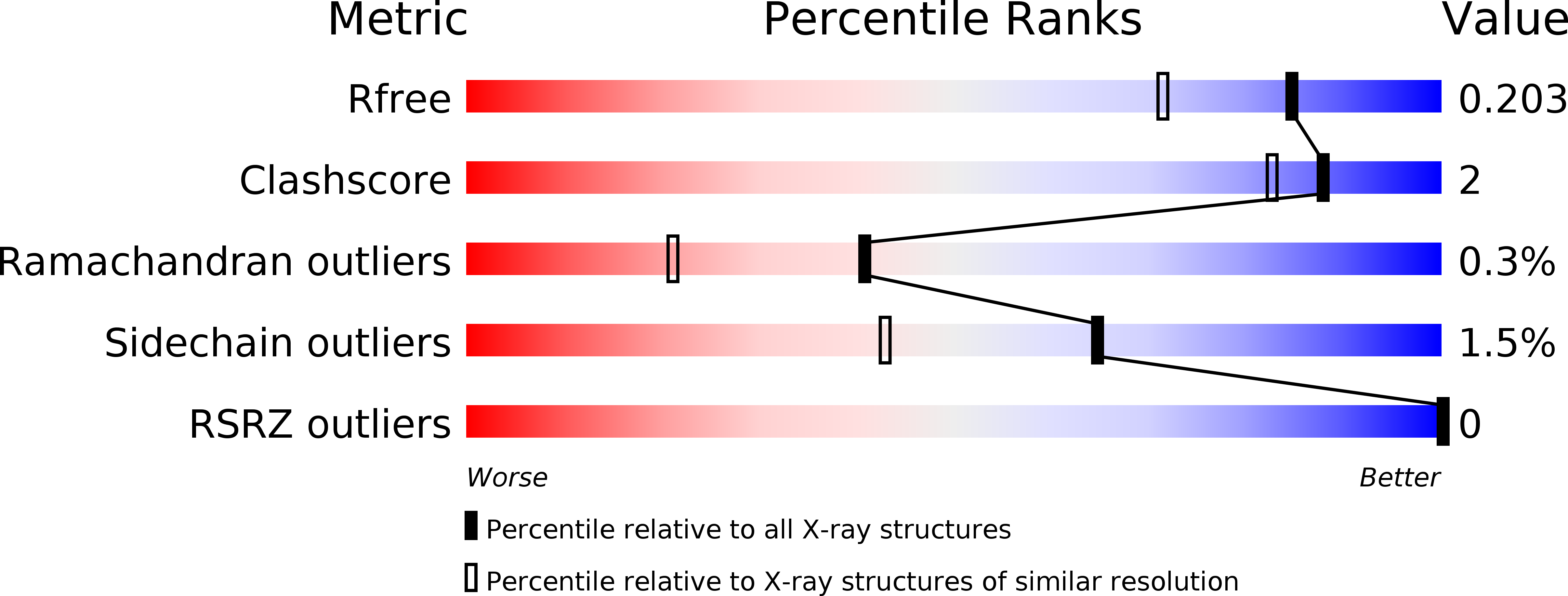
Desolvation of the substrate-binding protein TauA dictates ligand specificity for the alkanesulfonate ABC importer TauABC.
Qu, F., ElOmari, K., Wagner, A., De Simone, A., Beis, K.(2019) Biochem J 476: 3649-3660
- PubMed: 31802112
- DOI: https://doi.org/10.1042/BCJ20190779
- Primary Citation of Related Structures:
6SSY, 6ST0, 6ST1, 6STL - PubMed Abstract:
Under limiting sulfur availability, bacteria can assimilate sulfur from alkanesulfonates. Bacteria utilize ATP-binding cassette (ABC) transporters to internalise them for further processing to release sulfur. In gram-negative bacteria the TauABC and SsuABC ensure internalization, although, these two systems have common substrates, the former has been characterized as a taurine specific system. TauA and SsuA are substrate-binding proteins (SBPs) that bind and bring the alkanesulfonates to the ABC importer for transport. Here, we have determined the crystal structure of TauA and have characterized its thermodynamic binding parameters by isothermal titration calorimetry in complex with taurine and different alkanesulfonates. Our structures revealed that the coordination of the alkanesulfonates is conserved, with the exception of Asp205 that is absent from SsuA, but the thermodynamic parameters revealed a very high enthalpic penalty cost for binding of the other alkanesulfonates relative to taurine. Our molecular dynamic simulations indicated that the different levels of hydration of the binding site contributed to the selectivity for taurine over the other alkanesulfonates. Such selectivity mechanism is very likely to be employed by other SBPs of ABC transporters.
Organizational Affiliation:
Department of Life Sciences, Imperial College London, South Kensington, London SW7 2AZ, U.K.