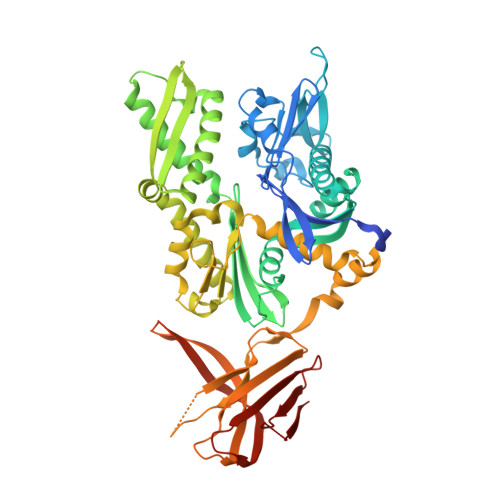
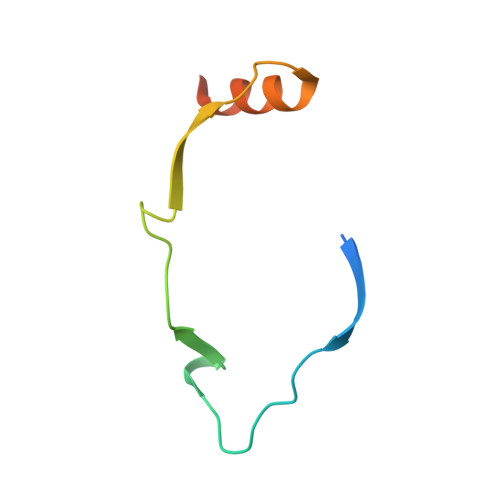
The ribosome-associated complex RAC serves in a relay that directs nascent chains to Ssb.
Zhang, Y., Valentin Gese, G., Conz, C., Lapouge, K., Kopp, J., Wolfle, T., Rospert, S., Sinning, I.(2020) Nat Commun 11: 1504-1504
- PubMed: 32198371
- DOI: https://doi.org/10.1038/s41467-020-15313-w
- Primary Citation of Related Structures:
6SR6 - PubMed Abstract:
The conserved ribosome-associated complex (RAC) consisting of Zuo1 (Hsp40) and Ssz1 (non-canonical Hsp70) acts together with the ribosome-bound Hsp70 chaperone Ssb in de novo protein folding at the ribosomal tunnel exit. Current models suggest that the function of Ssz1 is confined to the support of Zuo1, however, it is not known whether RAC by itself serves as a chaperone for nascent chains. Here we show that, via its rudimentary substrate binding domain (SBD), Ssz1 directly binds to emerging nascent chains prior to Ssb. Structural and biochemical analyses identify a conserved LP-motif at the Zuo1 N-terminus forming a polyproline-II helix, which binds to the Ssz1-SBD as a pseudo-substrate. The LP-motif competes with nascent chain binding to the Ssz1-SBD and modulates nascent chain transfer. The combined data indicate that Ssz1 is an active chaperone optimized for transient, low-affinity substrate binding, which ensures the flux of nascent chains through RAC/Ssb.
Organizational Affiliation:
Institute of Biochemistry and Molecular Biology, ZBMZ, Faculty of Medicine, University of Freiburg, D-79104 Freiburg, Germany; BIOSS Centre for Biological Signaling Studies, University of Freiburg, D-79104, Freiburg, Germany.