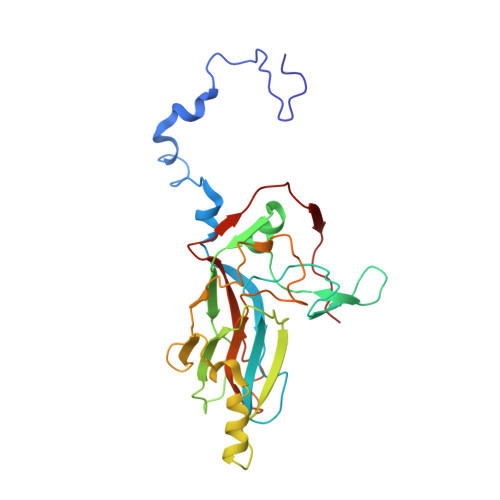
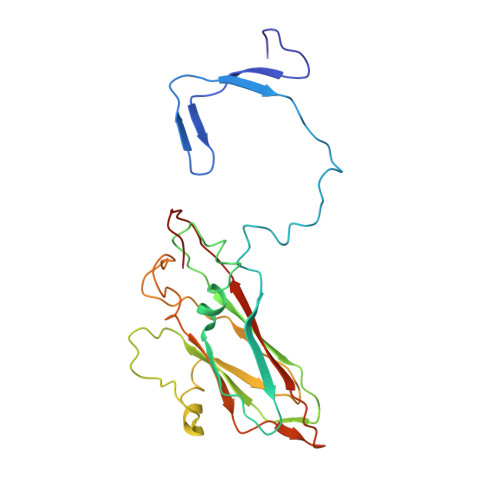
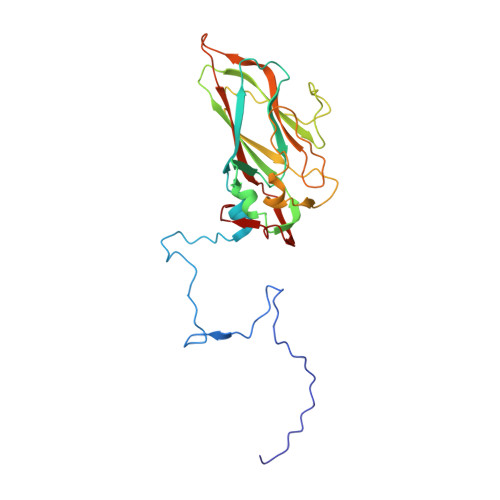
Capsid Structure of a Marine Algal Virus of the Order Picornavirales .
Munke, A., Kimura, K., Tomaru, Y., Okamoto, K.(2020) J Virol 94
- PubMed: 32024776
- DOI: https://doi.org/10.1128/JVI.01855-19
- Primary Citation of Related Structures:
6SHL - PubMed Abstract:
The order Picornavirales includes viruses that infect different kinds of eukaryotes and that share similar properties. The capsid proteins (CPs) of viruses in the order that infect unicellular organisms, such as algae, presumably possess certain characteristics that have changed little over the course of evolution, and thus these viruses may resemble the Picornavirales ancestor in some respects. Herein, we present the capsid structure of Chaetoceros tenuissimus RNA virus type II (CtenRNAV-II) determined using cryo-electron microscopy at a resolution of 3.1 Å, the first alga virus belonging to the family Marnaviridae of the order Picornavirales A structural comparison to related invertebrate and vertebrate viruses revealed a unique surface loop of the major CP VP1 that had not been observed previously, and further, revealed that another VP1 loop obscures the so-called canyon, which is a host-receptor binding site for many of the mammalian Picornavirales viruses. VP2 has an N-terminal tail, which has previously been reported as a primordial feature of Picornavirales viruses. The above-mentioned and other critical structural features provide new insights on three long-standing theories about Picornavirales : (i) the canyon hypothesis, (ii) the primordial VP2 domain swap, and (iii) the hypothesis that alga Picornavirales viruses could share characteristics with the Picornavirales ancestor. IMPORTANCE Identifying the acquired structural traits in virus capsids is important for elucidating what functions are essential among viruses that infect different hosts. The Picornavirales viruses infect a broad spectrum of hosts, ranging from unicellular algae to insects and mammals and include many human pathogens. Those viruses that infect unicellular protists, such as algae, are likely to have undergone fewer structural changes during the course of evolution compared to those viruses that infect multicellular eukaryotes and thus still share some characteristics with the Picornavirales ancestor. This article describes the first atomic capsid structure of an alga Marnavirus , CtenRNAV-II. A comparison to capsid structures of the related invertebrate and vertebrate viruses identified a number of structural traits that have been functionally acquired or lost during the course of evolution. These observations provide new insights on past theories on the viability and evolution of Picornavirales viruses.
Organizational Affiliation:
The Laboratory of Molecular Biophysics, Department of Cell and Molecular Biology, Uppsala University, Uppsala, Sweden anna.munke@icm.uu.se kenta.okamoto@icm.uu.se.