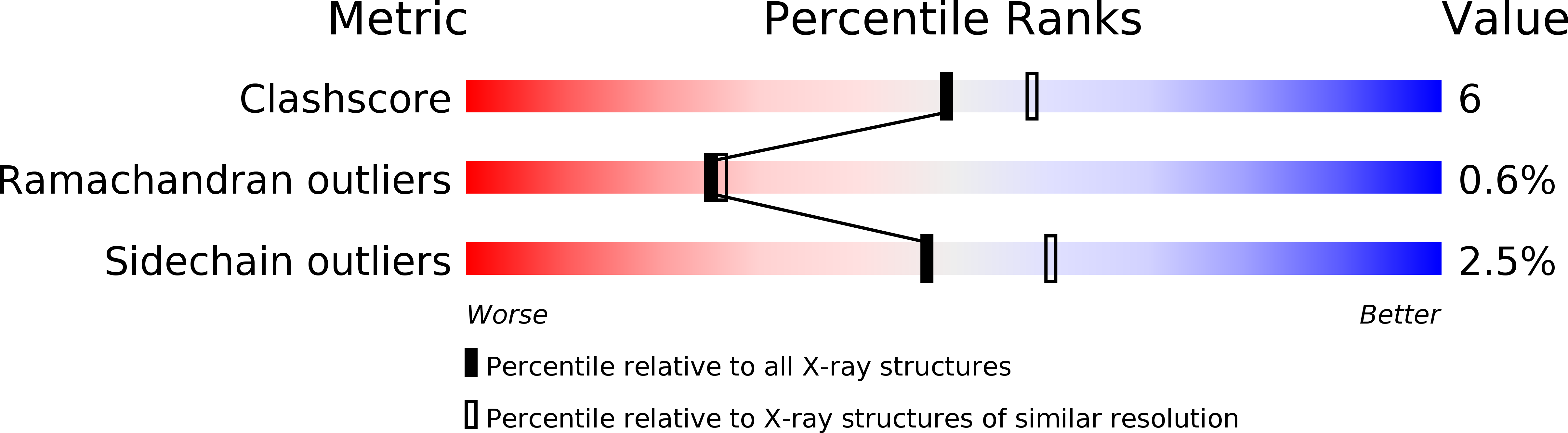Structure and Function of an Elongation Factor P Subfamily in Actinobacteria.
Pinheiro, B., Scheidler, C.M., Kielkowski, P., Schmid, M., Forne, I., Ye, S., Reiling, N., Takano, E., Imhof, A., Sieber, S.A., Schneider, S., Jung, K.(2020) Cell Rep 30: 4332-4342.e5
- PubMed: 32234471
- DOI: https://doi.org/10.1016/j.celrep.2020.03.009
- Primary Citation of Related Structures:
6S8Z - PubMed Abstract:
Translation of consecutive proline motifs causes ribosome stalling and requires rescue via the action of a specific translation elongation factor, EF-P in bacteria and archaeal/eukaryotic a/eIF5A. In Eukarya, Archaea, and all bacteria investigated so far, the functionality of this translation elongation factor depends on specific and rather unusual post-translational modifications. The phylum Actinobacteria, which includes the genera Corynebacterium, Mycobacterium, and Streptomyces, is of both medical and economic significance. Here, we report that EF-P is required in these bacteria in particular for the translation of proteins involved in amino acid and secondary metabolite production. Notably, EF-P of Actinobacteria species does not need any post-translational modification for activation. While the function and overall 3D structure of this EF-P type is conserved, the loop containing the conserved lysine is flanked by two essential prolines that rigidify it. Actinobacteria's EF-P represents a unique subfamily that works without any modification.
Organizational Affiliation:
Department of Biology I, Microbiology, Ludwig-Maximilians-Universität München, Martinsried, Germany.