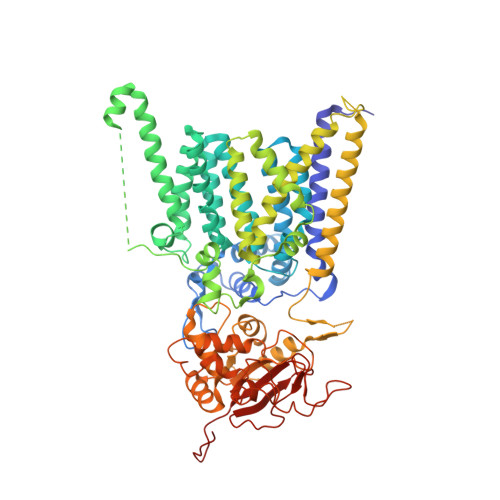
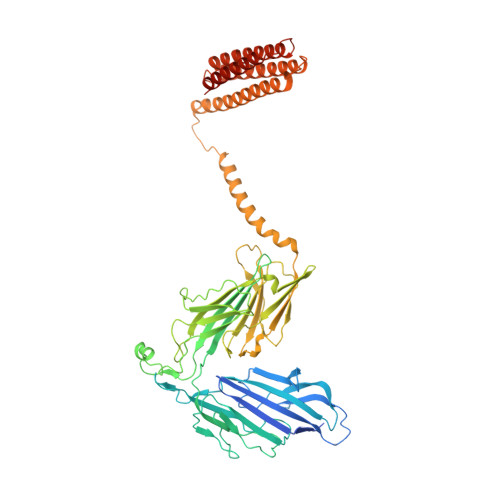
Cryo-electron microscopy structures of human oligosaccharyltransferase complexes OST-A and OST-B.
Ramirez, A.S., Kowal, J., Locher, K.P.(2019) Science 366: 1372-1375
- PubMed: 31831667
- DOI: https://doi.org/10.1126/science.aaz3505
- Primary Citation of Related Structures:
6S7O, 6S7T - PubMed Abstract:
Oligosaccharyltransferase (OST) catalyzes the transfer of a high-mannose glycan onto secretory proteins in the endoplasmic reticulum. Mammals express two distinct OST complexes that act in a cotranslational (OST-A) or posttranslocational (OST-B) manner. Here, we present high-resolution cryo-electron microscopy structures of human OST-A and OST-B. Although they have similar overall architectures, structural differences in the catalytic subunits STT3A and STT3B facilitate contacts to distinct OST subunits, DC2 in OST-A and MAGT1 in OST-B. In OST-A, interactions with TMEM258 and STT3A allow ribophorin-I to form a four-helix bundle that can bind to a translating ribosome, whereas the equivalent region is disordered in OST-B. We observed an acceptor peptide and dolichylphosphate bound to STT3B, but only dolichylphosphate in STT3A, suggesting distinct affinities of the two OST complexes for protein substrates.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, Eidgenössische Technische Hochschule (ETH), CH-8093 Zürich, Switzerland.