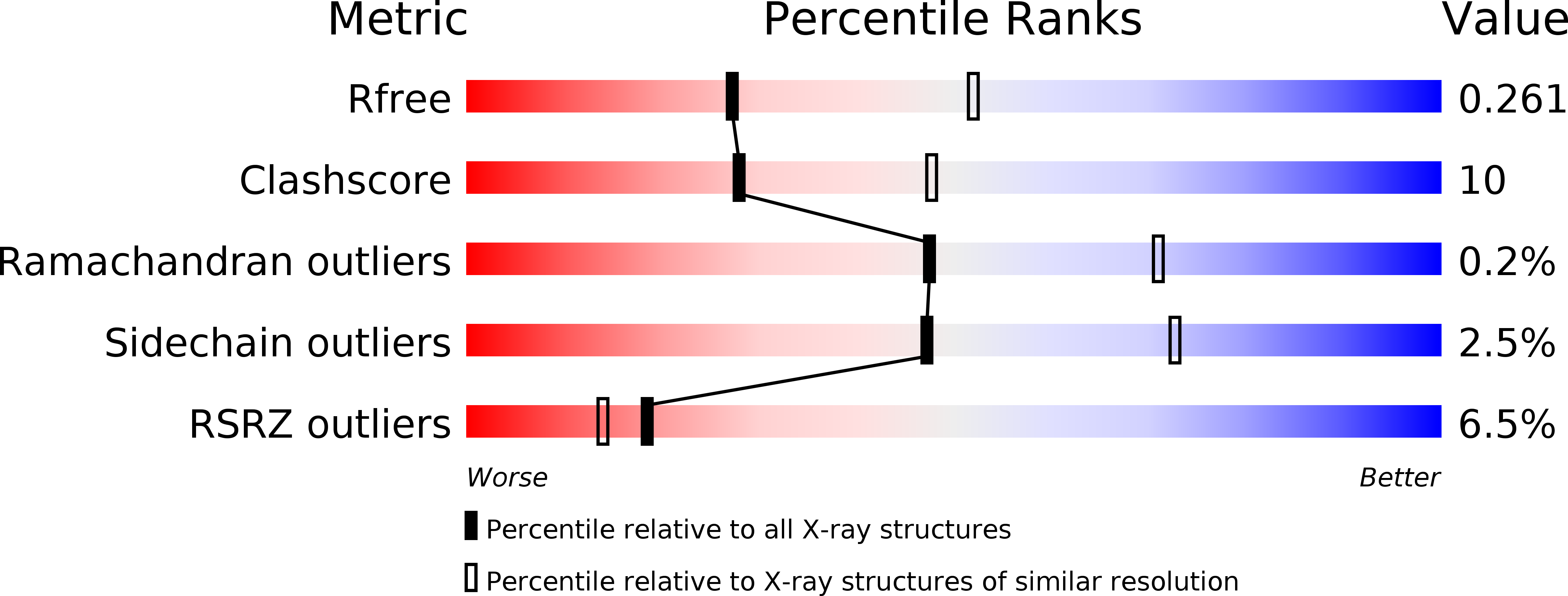
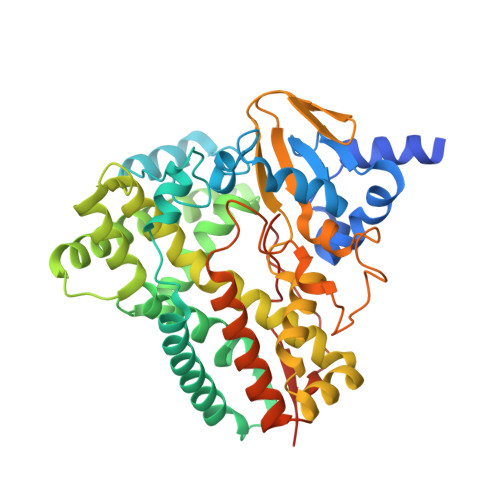
Crystal structure of bacterial CYP116B5 heme domain: New insights on class VII P450s structural flexibility and peroxygenase activity.
Ciaramella, A., Catucci, G., Gilardi, G., Di Nardo, G.(2019) Int J Biol Macromol 140: 577-587
- PubMed: 31430491
- DOI: https://doi.org/10.1016/j.ijbiomac.2019.08.141
- Primary Citation of Related Structures:
6RO8 - PubMed Abstract:
Class VII cytochromes P450 are self-sufficient enzymes carrying a phthalate family oxygenase-like reductase domain and a P450 domain fused in a single polypeptide chain. The biocatalytic applications of CYP116B members are limited by the need of the NADPH cofactor and the lack of crystal structures as a starting point for protein engineering. Nevertheless, we demonstrated that the heme domain of CYP116B5 can use hydrogen peroxide as electron donor bypassing the need of NADPH. Here, we report the crystal structure of CYP116B5 heme domain in complex with histidine at 2.6 Å of resolution. The structure reveals the typical P450 fold and a closed conformation with an active site cavity of 284 Å 3 in volume, accommodating a histidine molecule forming a hydrogen bond with the water molecule present as 6th heme iron ligand. MD simulations in the absence of any ligand revealed the opening of a tunnel connecting the active site to the protein surface through the movement of F-, G- and H-helices. A structural alignment with bacterial cytochromes P450 allowed the identification of amino acids in the proximal heme site potentially involved in peroxygenase activity. The availability of the crystal structure provides the bases for the structure-guided design of new biocatalysts.
Organizational Affiliation:
Department of Life Sciences and Systems Biology, University of Torino, Via Accademia Albertina 13, Torino, Italy.