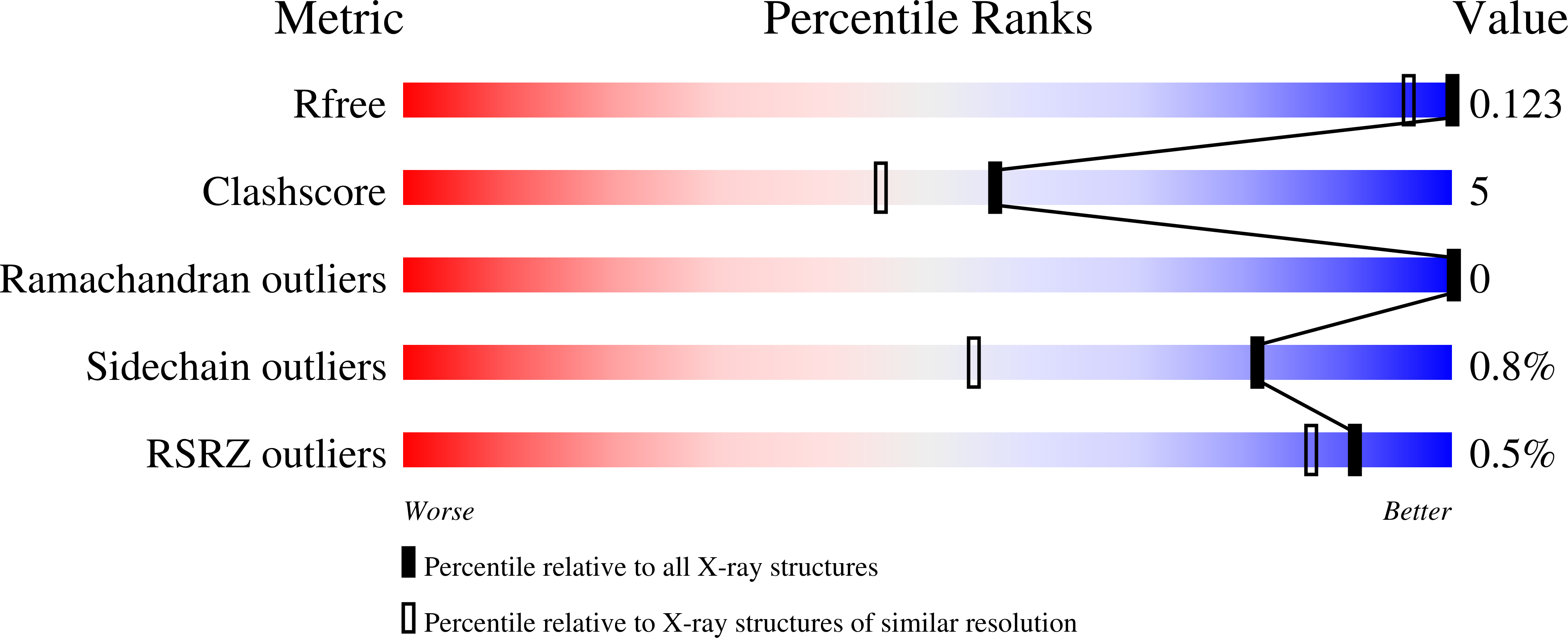
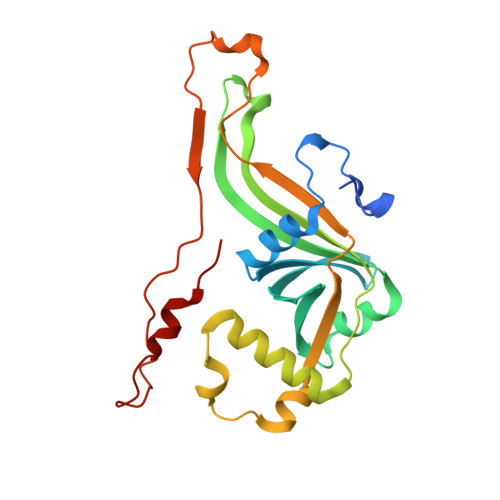
A low-potential terminal oxidase associated with the iron-only nitrogenase from the nitrogen-fixing bacteriumAzotobacter vinelandii.
Varghese, F., Kabasakal, B.V., Cotton, C.A.R., Schumacher, J., Rutherford, A.W., Fantuzzi, A., Murray, J.W.(2019) J Biol Chem 294: 9367-9376
- PubMed: 31043481
- DOI: https://doi.org/10.1074/jbc.RA118.007285
- Primary Citation of Related Structures:
6RK0 - PubMed Abstract:
The biological route for nitrogen gas entering the biosphere is reduction to ammonia by the nitrogenase enzyme, which is inactivated by oxygen. Three types of nitrogenase exist, the least-studied of which is the iron-only nitrogenase. The Anf3 protein in the bacterium Rhodobacter capsulatus is essential for diazotrophic ( i.e. nitrogen-fixing) growth with the iron-only nitrogenase, but its enzymatic activity and function are unknown. Here, we biochemically and structurally characterize Anf3 from the model diazotrophic bacterium Azotobacter vinelandii Determining the Anf3 crystal structure to atomic resolution, we observed that it is a dimeric flavocytochrome with an unusually close interaction between the heme and the FAD cofactors. Measuring the reduction potentials by spectroelectrochemical redox titration, we observed values of -420 ± 10 and -330 ± 10 mV for the two FAD potentials and -340 ± 1 mV for the heme. We further show that Anf3 accepts electrons from spinach ferredoxin and that Anf3 consumes oxygen without generating superoxide or hydrogen peroxide. We predict that Anf3 protects the iron-only nitrogenase from oxygen inactivation by functioning as an oxidase in respiratory protection, with flavodoxin or ferredoxin as the physiological electron donors.
Organizational Affiliation:
From the Department of Life Sciences, Imperial College London, London SW7 2AZ, United Kingdom.