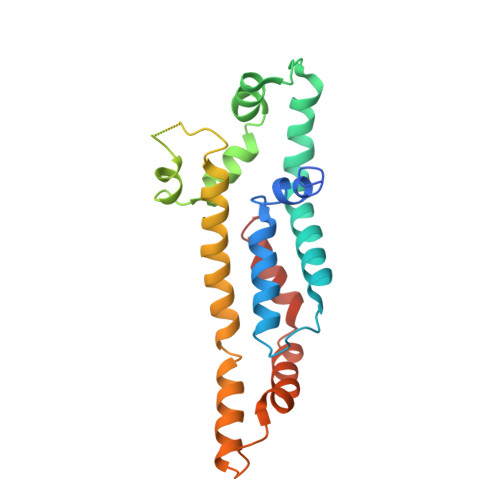
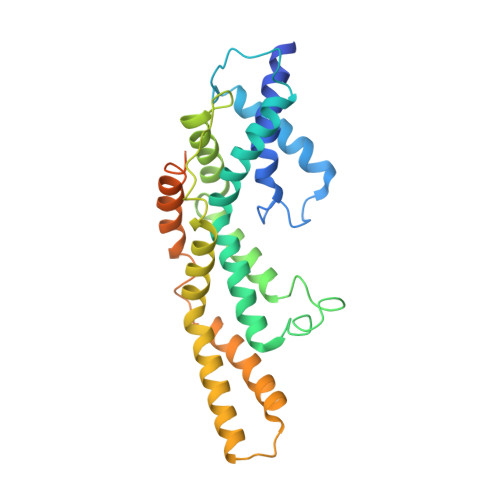
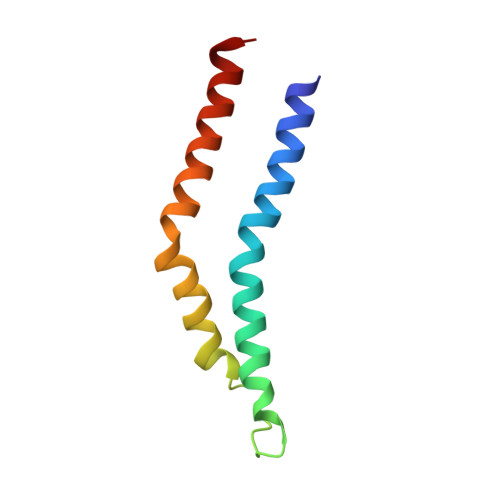
The Structure of an Injectisome Export Gate Demonstrates Conservation of Architecture in the Core Export Gate between Flagellar and Virulence Type III Secretion Systems.
Johnson, S., Kuhlen, L., Deme, J.C., Abrusci, P., Lea, S.M.(2019) mBio 10
- PubMed: 31239376
- DOI: https://doi.org/10.1128/mBio.00818-19
- Primary Citation of Related Structures:
6R69, 6R6B - PubMed Abstract:
Export of proteins through type III secretion systems (T3SS) is critical for motility and virulence of many major bacterial pathogens. Proteins are exported through a genetically defined export gate complex consisting of three proteins. We have recently shown at 4.2 Å that the flagellar complex of these three putative membrane proteins (FliPQR in flagellar systems, SctRST in virulence systems) assembles into an extramembrane helical assembly that likely seeds correct assembly of the rod. Here we present the structure of an equivalent complex from the Shigella virulence system at 3.5 Å by cryo-electron microscopy. This higher-resolution structure yields a more precise description of the structure and confirms the prediction of structural conservation in this core complex. Analysis of particle heterogeneity also suggests how the SctS/FliQ subunits sequentially assemble in the complex. IMPORTANCE Although predicted on the basis of sequence conservation, the work presented here formally demonstrates that all classes of type III secretion systems, flagellar or virulence, share the same architecture at the level of the core structures. This absolute conservation of the unusual extramembrane structure of the core export gate complex now allows work to move to focusing on both mechanistic studies of type III but also on fundamental studies of how such a complex is assembled.
Organizational Affiliation:
Sir William Dunn School of Pathology, University of Oxford, Oxford, United Kingdom.