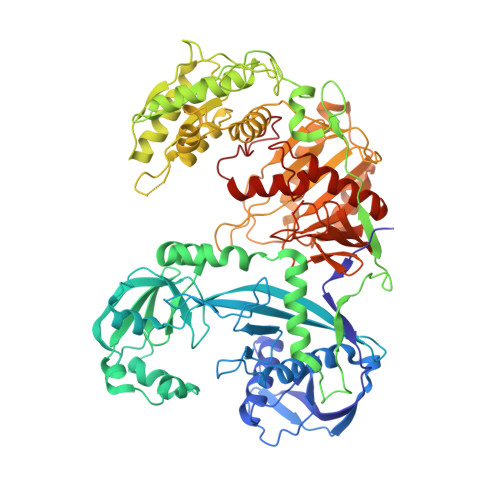
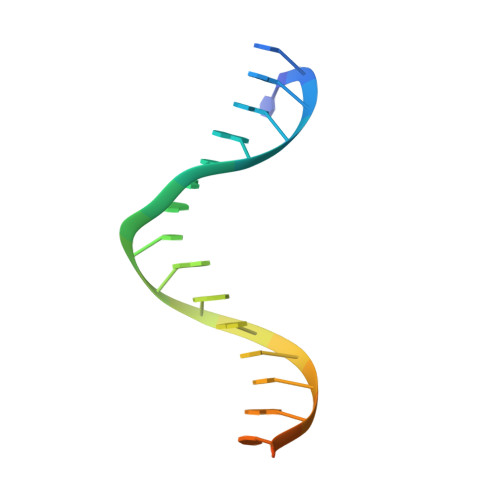
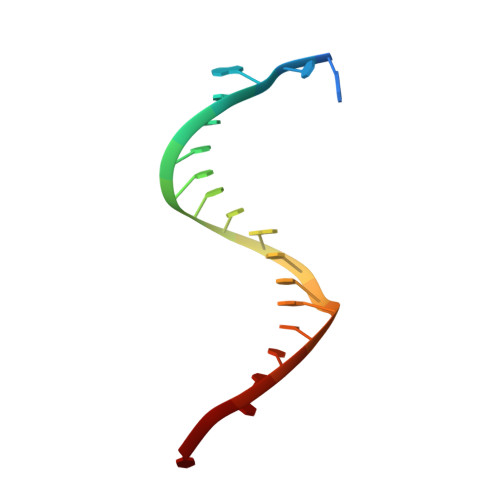
DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute.
Hegge, J.W., Swarts, D.C., Chandradoss, S.D., Cui, T.J., Kneppers, J., Jinek, M., Joo, C., van der Oost, J.(2019) Nucleic Acids Res 47: 5809-5821
- PubMed: 31069393
- DOI: https://doi.org/10.1093/nar/gkz306
- Primary Citation of Related Structures:
6QZK - PubMed Abstract:
Prokaryotic Argonaute proteins (pAgos) constitute a diverse group of endonucleases of which some mediate host defense by utilizing small interfering DNA guides (siDNA) to cleave complementary invading DNA. This activity can be repurposed for programmable DNA cleavage. However, currently characterized DNA-cleaving pAgos require elevated temperatures (≥65°C) for their activity, making them less suitable for applications that require moderate temperatures, such as genome editing. Here, we report the functional and structural characterization of the siDNA-guided DNA-targeting pAgo from the mesophilic bacterium Clostridium butyricum (CbAgo). CbAgo displays a preference for siDNAs that have a deoxyadenosine at the 5'-end and thymidines at nucleotides 2-4. Furthermore, CbAgo mediates DNA-guided DNA cleavage of AT-rich double stranded DNA at moderate temperatures (37°C). This study demonstrates that certain pAgos are capable of programmable DNA cleavage at moderate temperatures and thereby expands the scope of the potential pAgo-based applications.
Organizational Affiliation:
Laboratory of Microbiology, Department of Agrotechnology and Food Sciences, Wageningen University, 6708 WE Wageningen, The Netherlands.