Mechanistic and Crystallographic Studies of Azoreductase AzoA fromBacillus wakoensisA01.
Romero, E., Savino, S., Fraaije, M.W., Loncar, N.(2020) ACS Chem Biol 15: 504-512
- PubMed: 31967777
- DOI: https://doi.org/10.1021/acschembio.9b00970
- Primary Citation of Related Structures:
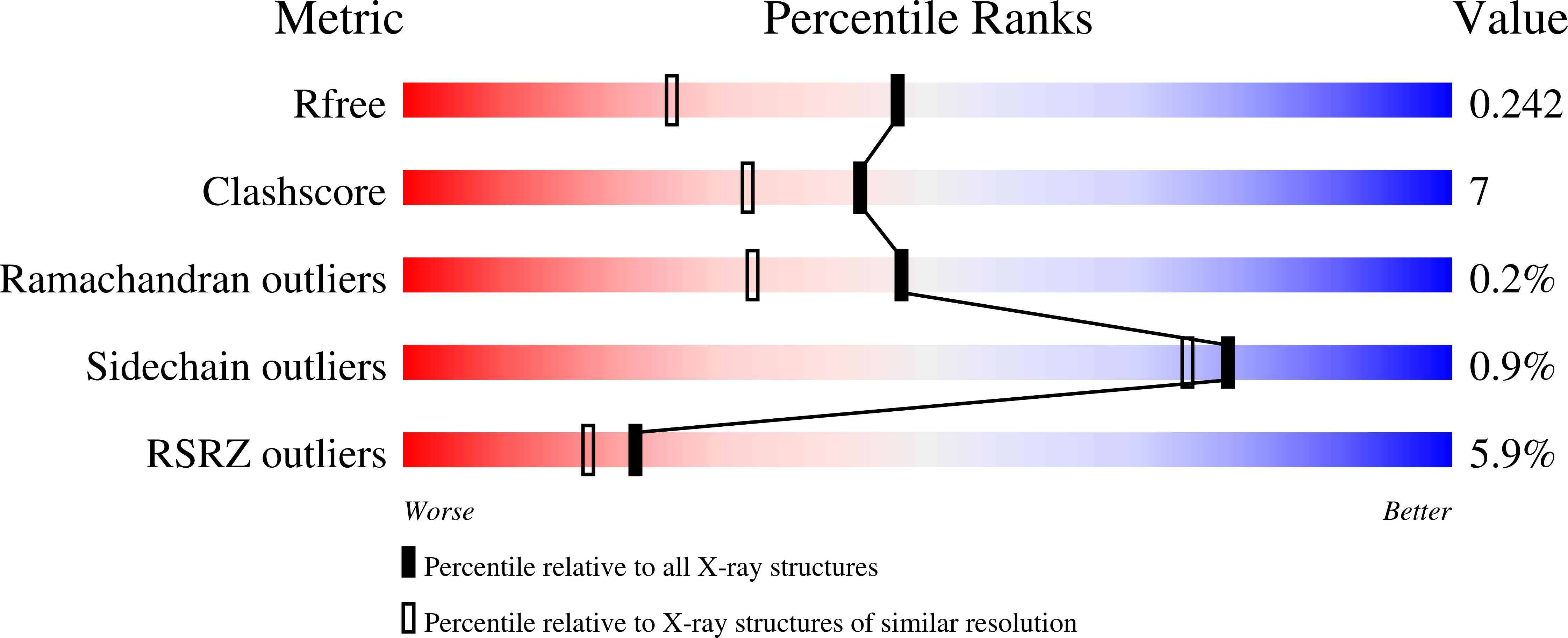
6QU0 - PubMed Abstract:
The azoreductase AzoA from the alkali-tolerant Bacillus wakoensis A01 has been studied to reveal its structural and mechanistic details. For this, a recombinant expression system was developed which yields impressive amounts of fully active enzyme. The purified holo enzyme is remarkably solvent-tolerant and thermostable with an apparent melting temperature of 71 °C. The dimeric enzyme contains FMN as a prosthetic group and is strictly NADH dependent. While AzoA shows a negligible ability to use molecular oxygen as an electron acceptor, it is efficient in reducing various azo dyes and quinones. The kinetic and catalytic mechanism has been studied in detail using steady state kinetic analyses and stopped-flow studies. The data show that AzoA performs quinone and azo dye reductions via a two-electron transfer. Moreover, quinones were shown to be much better substrates ( k cat values of 100-400 s -1 for several naphtoquinones) when compared with azo dyes. This suggests that the physiological role of AzoA and sequence-related microbial reductases is linked to quinone reductions and that they can better be annotated as quinone reductases. The structure of AzoA has been determined in complex with FMN at 1.8 Å resolution. AzoA displays unique features in the active site providing clues for explaining its catalytic and thermostability features. An uncommon loop, when compared with sequence-related reductases, forms an active site lid with Trp60 acting as an anchor. Several Trp60 mutants have been analyzed disclosing an important role of this residue in the stability of AzoA, while they retained activity. Structural details are discussed in relation to other azo and quinone reductases. This study provides new insights into the molecular functioning of AzoA and sequence-related reductases.
Organizational Affiliation:
Molecular Enzymology Group , University of Groningen , Nijenborgh 4 , 9747AG Groningen , The Netherlands.