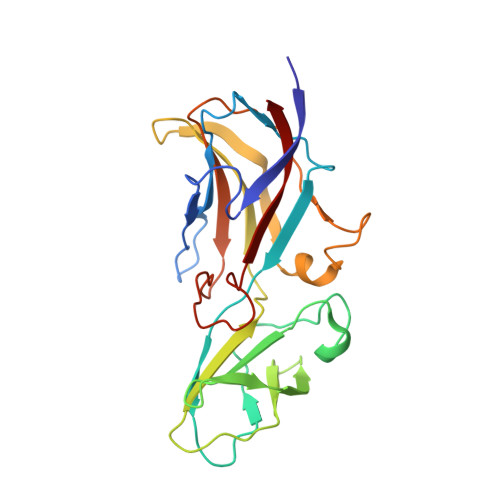
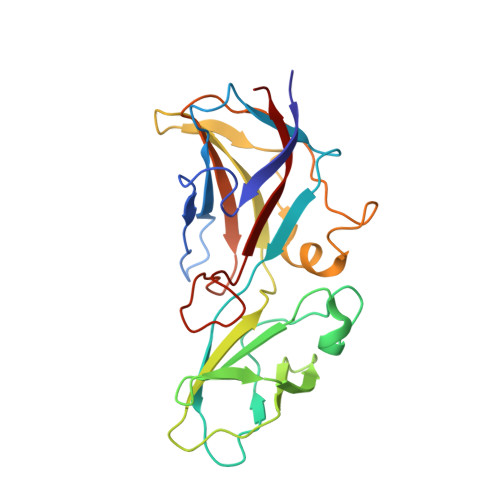
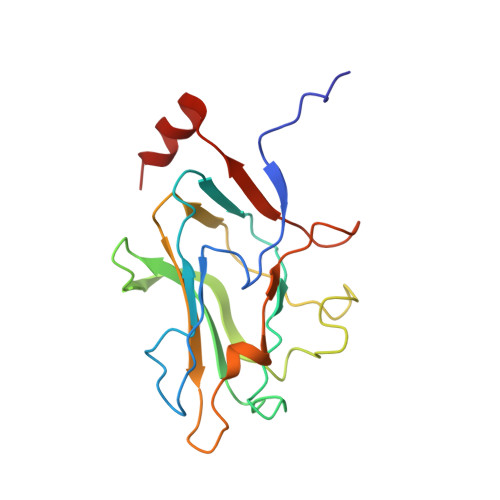
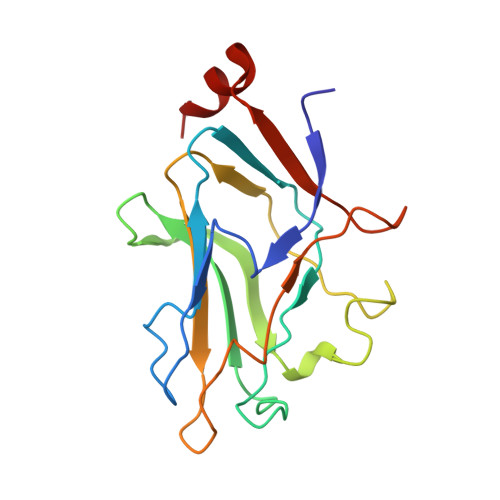
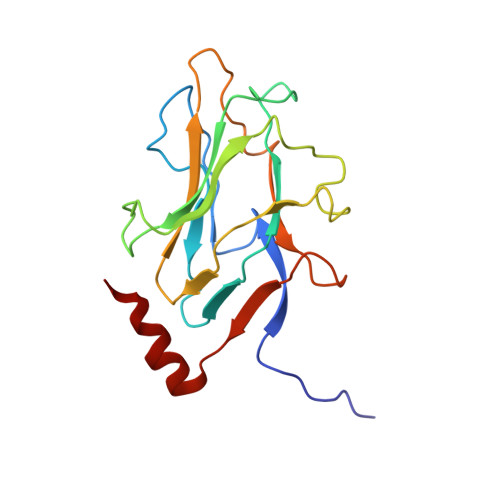
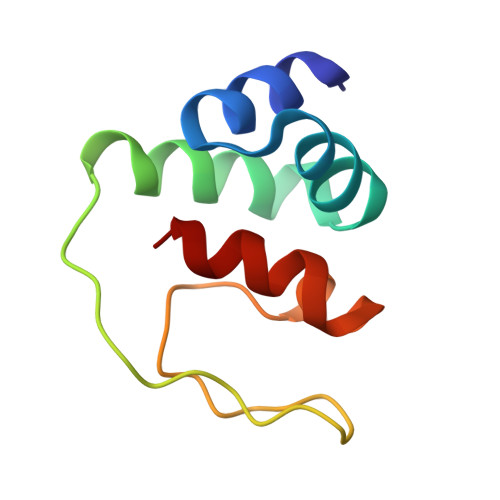
Assembly of complex viruses exemplified by a halophilic euryarchaeal virus.
Colibus, L., Roine, E., Walter, T.S., Ilca, S.L., Wang, X., Wang, N., Roseman, A.M., Bamford, D., Huiskonen, J.T., Stuart, D.I.(2019) Nat Commun 10: 1456-1456
- PubMed: 30926810
- DOI: https://doi.org/10.1038/s41467-019-09451-z
- Primary Citation of Related Structures:
6QT9 - PubMed Abstract:
Many of the largest known viruses belong to the PRD1-adeno structural lineage characterised by conserved pseudo-hexameric capsomers composed of three copies of a single major capsid protein (MCP). Here, by high-resolution cryo-EM analysis, we show that a class of archaeal viruses possess hetero-hexameric MCPs which mimic the PRD1-adeno lineage trimer. These hetero-hexamers are built from heterodimers and utilise a jigsaw-puzzle system of pegs and holes, and underlying minor capsid proteins, to assemble the capsid laterally from the 5-fold vertices. At these vertices proteins engage inwards with the internal membrane vesicle whilst 2-fold symmetric horn-like structures protrude outwards. The horns are assembled from repeated globular domains attached to a central spine, presumably facilitating multimeric attachment to the cell receptor. Such viruses may represent precursors of the main PRD1-adeno lineage, similarly engaging cell-receptors via 5-fold spikes and using minor proteins to define particle size.
Organizational Affiliation:
Division of Structural Biology, University of Oxford, Wellcome Centre for Human Genetics, Oxford, OX3 7BN, UK.