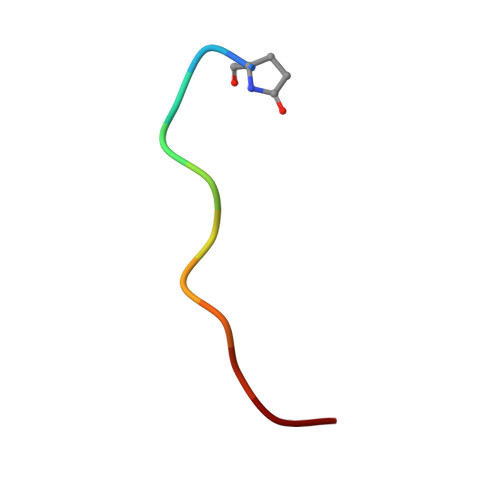
Structural basis for the C-domain-selective angiotensin-converting enzyme inhibition by bradykinin-potentiating peptide b (BPPb).
Sturrock, E.D., Lubbe, L., Cozier, G.E., Schwager, S.L.U., Arowolo, A.T., Arendse, L.B., Belcher, E., Acharya, K.R.(2019) Biochem J 476: 1553-1570
- PubMed: 31072910
- DOI: https://doi.org/10.1042/BCJ20190290
- Primary Citation of Related Structures:
6QS1 - PubMed Abstract:
Angiotensin-converting enzyme (ACE) is a zinc metalloprotease best known for its role in blood pressure regulation. ACE consists of two homologous catalytic domains, the N- and C-domain, that display distinct but overlapping catalytic functions in vivo owing to subtle differences in substrate specificity. While current generation ACE inhibitors target both ACE domains, domain-selective ACE inhibitors may be clinically advantageous, either reducing side effects or having utility in new indications. Here, we used site-directed mutagenesis, an ACE chimera and X-ray crystallography to unveil the molecular basis for C-domain-selective ACE inhibition by the bradykinin-potentiating peptide b (BPPb), naturally present in Brazilian pit viper venom. We present the BPPb N-domain structure in comparison with the previously reported BPPb C-domain structure and highlight key differences in peptide interactions with the S 4 to S 9 subsites. This suggests the involvement of these subsites in conferring C-domain-selective BPPb binding, in agreement with the mutagenesis results where unique residues governing differences in active site exposure, lid structure and dynamics between the two domains were the major drivers for C-domain-selective BPPb binding. Mere disruption of BPPb interactions with unique S 2 and S 4 subsite residues, which synergistically assist in BPPb binding, was insufficient to abolish C-domain selectivity. The combination of unique S 9 -S 4 and S 2 ' subsite C-domain residues was required for the favourable entry, orientation and thus, selective binding of the peptide. This emphasizes the need to consider factors other than direct protein-inhibitor interactions to guide the design of domain-selective ACE inhibitors, especially in the case of larger peptides.
Organizational Affiliation:
Department of Integrative Biomedical Sciences, Institute of Infectious Disease and Molecular Medicine, University of Cape Town, Observatory, 7925, Cape Town, Republic of South Africa edward.sturrock@uct.ac.za.