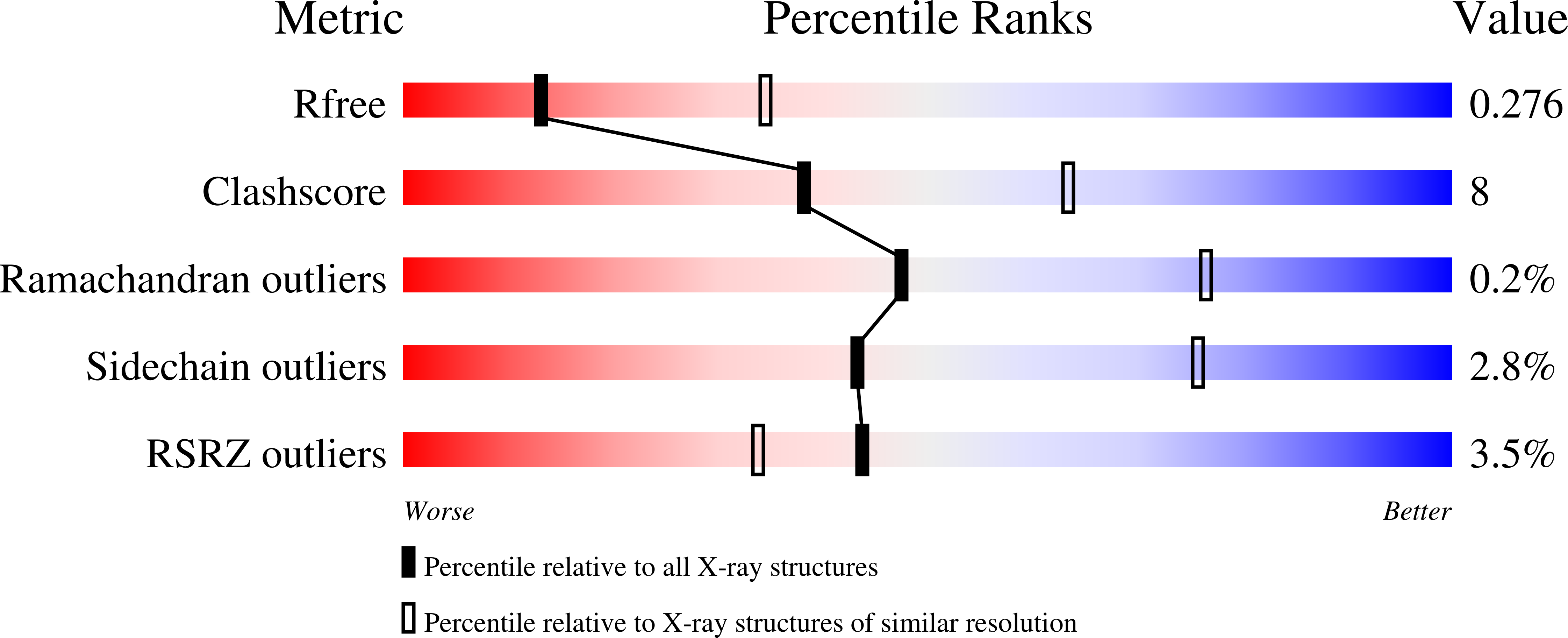
Mammalian-like type II glutaminyl cyclases in Porphyromonas gingivalis and other oral pathogenic bacteria as targets for treatment of periodontitis.
Taudte, N., Linnert, M., Rahfeld, J.U., Piechotta, A., Ramsbeck, D., Buchholz, M., Kolenko, P., Parthier, C., Houston, J.A., Veillard, F., Eick, S., Potempa, J., Schilling, S., Demuth, H.U., Stubbs, M.T.(2021) J Biol Chem 296: 100263-100263
- PubMed: 33837744
- DOI: https://doi.org/10.1016/j.jbc.2021.100263
- Primary Citation of Related Structures:
6QQL, 6QRO - PubMed Abstract:
The development of a targeted therapy would significantly improve the treatment of periodontitis and its associated diseases including Alzheimer's disease, rheumatoid arthritis, and cardiovascular diseases. Glutaminyl cyclases (QCs) from the oral pathogens Porphyromonas gingivalis, Tannerella forsythia, and Prevotella intermedia represent attractive target enzymes for small-molecule inhibitor development, as their action is likely to stabilize essential periplasmic and outer membrane proteins by N-terminal pyroglutamination. In contrast to other microbial QCs that utilize the so-called type I enzymes, these oral pathogens possess sequences corresponding to type II QCs, observed hitherto only in animals. However, whether differences between these bacteroidal QCs and animal QCs are sufficient to enable development of selective inhibitors is not clear. To learn more, we recombinantly expressed all three QCs. They exhibit comparable catalytic efficiencies and are inhibited by metal chelators. Crystal structures of the enzymes from P. gingivalis (PgQC) and T. forsythia (TfQC) reveal a tertiary structure composed of an eight-stranded β-sheet surrounded by seven α-helices, typical of animal type II QCs. In each case, an active site Zn ion is tetrahedrally coordinated by conserved residues. Nevertheless, significant differences to mammalian enzymes are found around the active site of the bacteroidal enzymes. Application of a PgQC-selective inhibitor described here for the first time results in growth inhibition of two P. gingivalis clinical isolates in a dose-dependent manner. The insights gained by these studies will assist in the development of highly specific small-molecule bacteroidal QC inhibitors, paving the way for alternative therapies against periodontitis and associated diseases.
Organizational Affiliation:
Periotrap Pharmaceuticals GmbH, Halle (Saale), Germany.