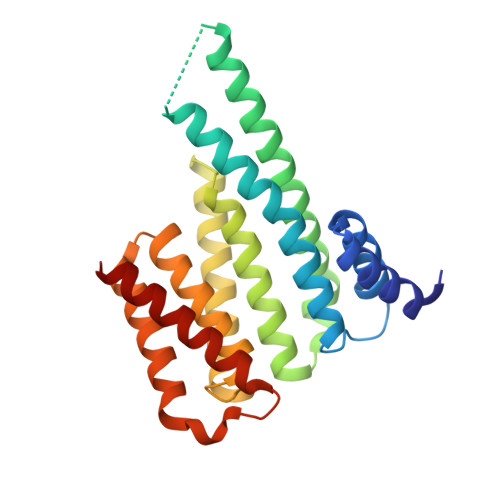
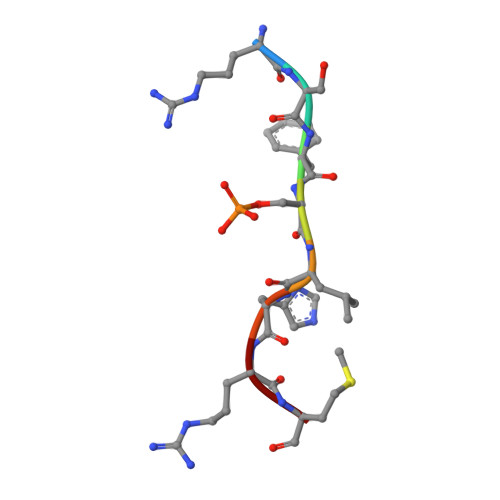
The activity of Saccharomyces cerevisiae Na+, K+/H+antiporter Nha1 is negatively regulated by 14-3-3 protein binding at serine 481.
Smidova, A., Stankova, K., Petrvalska, O., Lazar, J., Sychrova, H., Obsil, T., Zimmermannova, O., Obsilova, V.(2019) Biochim Biophys Acta Mol Cell Res 1866: 118534-118534
- PubMed: 31446061
- DOI: https://doi.org/10.1016/j.bbamcr.2019.118534
- Primary Citation of Related Structures:
6QK8 - PubMed Abstract:
Na + /H + antiporters are involved in ensuring optimal intracellular concentrations of alkali-metal cations and protons in most organisms. In Saccharomyces cerevisiae, the plasma-membrane Na + , K + /H + antiporter Nha1 mediates Na + and K + efflux, which is important for cell growth in the presence of salts. Nha1 belongs among housekeeping proteins and, due to its ability to export K + , it has many physiological functions. The Nha1 transport activity is regulated through its long, hydrophilic and unstructured C-terminus (554 of 985 aa). Although Nha1 has been previously shown to interact with the yeast 14-3-3 isoform (Bmh2), the binding site remains unknown. In this work, we identified the residues through which Nha1 interacts with the 14-3-3 protein. Biophysical characterization of the interaction between the C-terminal polypeptide of Nha1 and Bmh proteins in vitro revealed that the 14-3-3 protein binds to phosphorylated Ser481 of Nha1, and the crystal structure of the phosphopeptide containing Ser481 bound to Bmh1 provided the structural basis of this interaction. Our data indicate that 14-3-3 binding induces a disorder-to-order transition of the C-terminus of Nha1, and in vivo experiments showed that the mutation of Ser481 to Ala significantly increases cation efflux activity via Nha1, which renders cells sensitive to low K + concentrations. Hence, 14-3-3 binding is apparently essential for the negative regulation of Nha1 activity, which should be low under standard growth conditions, when low amounts of toxic salts are present and yeast cells need to accumulate high amounts of K + .
Organizational Affiliation:
Department of Structural Biology of Signaling Proteins, Division BIOCEV, Institute of Physiology of the Czech Academy of Sciences, 252 50 Vestec, Czech Republic.