Insights into the structure and function of Est3 from the Hansenula polymorpha telomerase.
Shepelev, N.M., Mariasina, S.S., Mantsyzov, A.B., Malyavko, A.N., Efimov, S.V., Petrova, O.A., Rodina, E.V., Zvereva, M.I., Dontsova, O.A., Polshakov, V.I.(2020) Sci Rep 10: 11109-11109
- PubMed: 32632130
- DOI: https://doi.org/10.1038/s41598-020-68107-x
- Primary Citation of Related Structures:
6Q44 - PubMed Abstract:
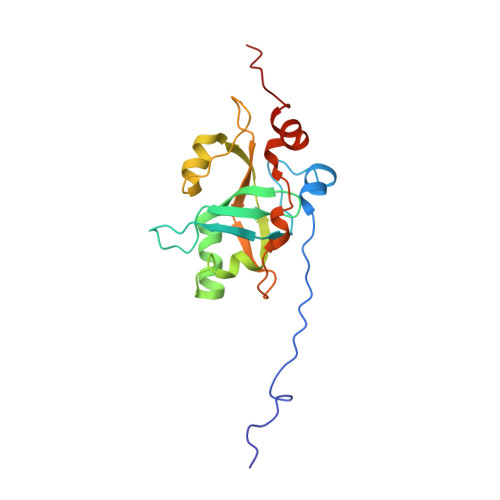
Telomerase is a ribonucleoprotein enzyme, which maintains genome integrity in eukaryotes and ensures continuous cellular proliferation. Telomerase holoenzyme from the thermotolerant yeast Hansenula polymorpha, in addition to the catalytic subunit (TERT) and telomerase RNA (TER), contains accessory proteins Est1 and Est3, which are essential for in vivo telomerase function. Here we report the high-resolution structure of Est3 from Hansenula polymorpha (HpEst3) in solution, as well as the characterization of its functional relationships with other components of telomerase. The overall structure of HpEst3 is similar to that of Est3 from Saccharomyces cerevisiae and human TPP1. We have shown that telomerase activity in H. polymorpha relies on both Est3 and Est1 proteins in a functionally symmetrical manner. The absence of either Est3 or Est1 prevents formation of a stable ribonucleoprotein complex, weakens binding of a second protein to TER, and decreases the amount of cellular TERT, presumably due to the destabilization of telomerase RNP. NMR probing has shown no direct in vitro interactions of free Est3 either with the N-terminal domain of TERT or with DNA or RNA fragments mimicking the probable telomerase environment. Our findings corroborate the idea that telomerase possesses the evolutionarily variable functionality within the conservative structural context.
Organizational Affiliation:
M.V. Lomonosov Moscow State University, Moscow, 119991, Russia.