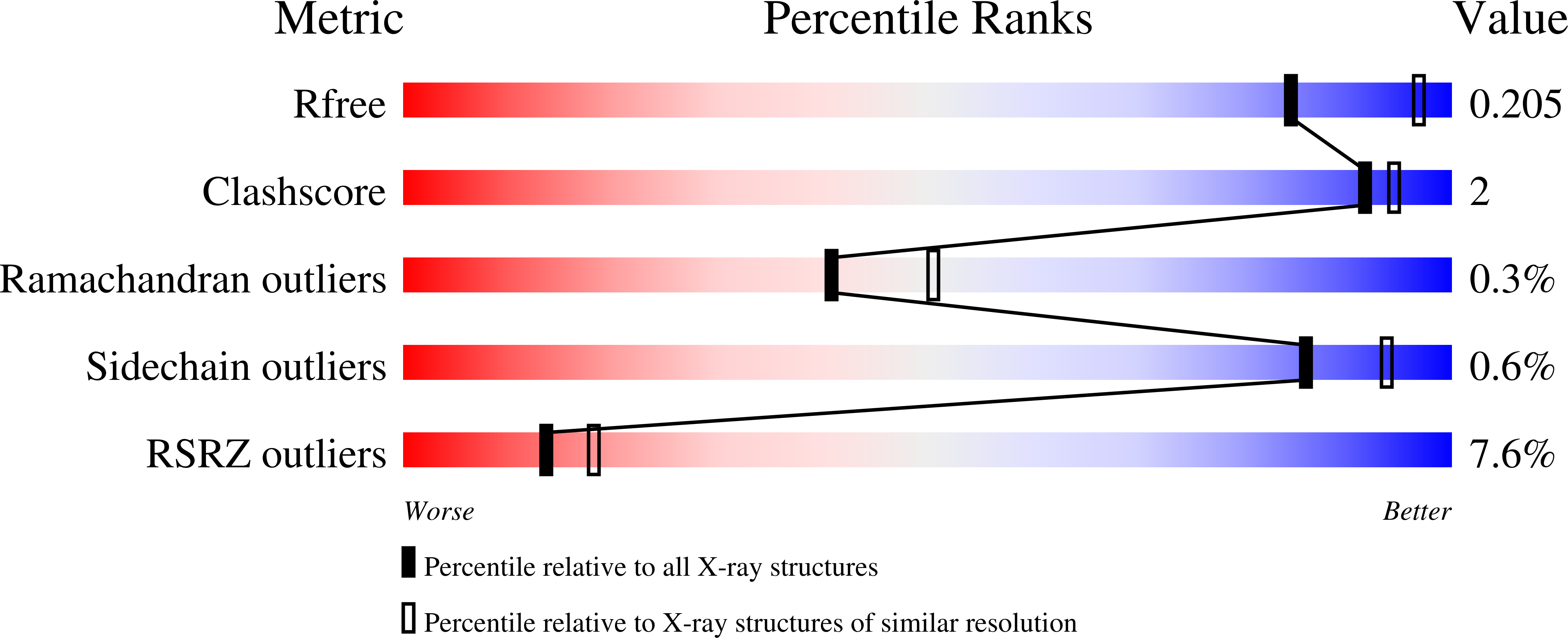
Crystal Structure of the Mannose-6-Phosphate Uncovering Enzyme.
Gorelik, A., Illes, K., Nagar, B.(2020) Structure 28: 426-436.e3
- PubMed: 32109365
- DOI: https://doi.org/10.1016/j.str.2020.02.001
- Primary Citation of Related Structures:
6PKG, 6PKH, 6PKI, 6PKU, 6PKY, 6U11 - PubMed Abstract:
Most lysosomal hydrolytic enzymes reach their destination via the mannose-6-phosphate (M6P) pathway. The enzyme N-acetylglucosamine-1-phosphodiester α-N-acetylglucosaminidase (NAGPA, or "uncovering enzyme") catalyzes the second step in the M6P tag formation, namely the removal of the masking N-acetylglucosamine (GlcNAc) portion. Defects in this protein are associated with non-syndromic stuttering. To gain a better understanding of the function and regulation of this enzyme, we determined its crystal structure. The propeptide binds in a groove on the globular catalytic domain, blocking active site access. High-affinity substrate binding is enabled by a conformational switch in an active site loop. The protein recognizes the GlcNAc and phosphate portions of its substrate, but not the mannose moiety of the glycan. Based on enzymatic and 1 H-NMR analysis, a catalytic mechanism is proposed. Crystallographic and solution scattering analyses suggest that the C-terminal domain forms a long flexible stem that extends the enzyme away from the Golgi membrane.
Organizational Affiliation:
Department of Biochemistry, McGill University, Room 464, 3649 Promenade Sir-William-Osler, Montreal, QC H3G 0B1, Canada.