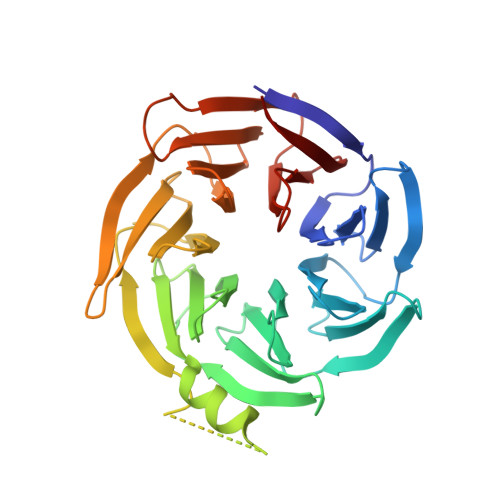
Crystal structure of W repeat domain of human coatomer subunit Alpha (COPA)
Halabelian, L., Zeng, H., Dong, A., Loppnau, P., Seitova, A., Hutchinson, A., Bountra, C., Edwards, A.M., Arrowsmith, C.H., Structural Genomics Consortium (SGC)To be published.