Structural Basis for a Species-Specific Determinant of an SIV Vif Protein toward Hominid APOBEC3G Antagonism.
Binning, J.M., Chesarino, N.M., Emerman, M., Gross, J.D.(2019) Cell Host Microbe 26: 739-747.e4
- PubMed: 31830442
- DOI: https://doi.org/10.1016/j.chom.2019.10.014
- Primary Citation of Related Structures:
6P59 - PubMed Abstract:
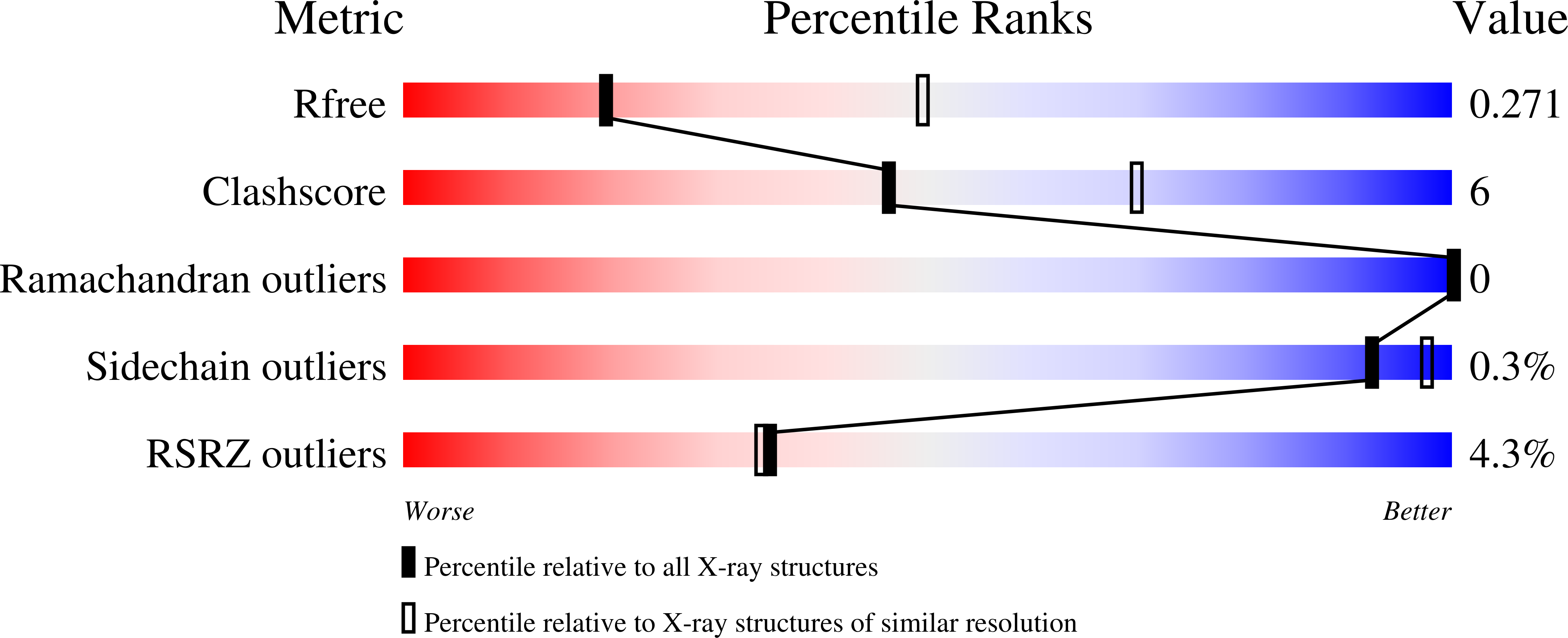
Primate lentiviruses encode a Vif protein that counteracts the host antiviral APOBEC3 (A3) family members. The adaptation of Vif to species-specific A3 determinants is a critical event that allowed the spillover of a lentivirus from monkey reservoirs to chimpanzees and subsequently to humans, which gave rise to HIV-1 and the acquired immune deficiency syndrome (AIDS) pandemic. How Vif-A3 protein interactions are remodeled during evolution is unclear. Here, we report a 2.94 Å crystal structure of the Vif substrate receptor complex from simian immunodeficiency virus isolated from red-capped mangabey (SIVrcm). The structure of the SIVrcm Vif complex illuminates the stage of lentiviral Vif evolution that is immediately prior to entering hominid primates. Structure-function studies reveal the adaptations that allowed SIVrcm Vif to antagonize hominid A3G. These studies show a partitioning between an evolutionarily dynamic specificity determinant and a conserved protein interacting surface on Vif that enables adaptation while maintaining protein interactions required for potent A3 antagonism.
Organizational Affiliation:
Department of Pharmaceutical Chemistry, University of California, San Francisco, San Francisco, CA 94158, USA.