Putative oxidoreductase from Escherichia coli str. K-12
Osipiuk, J., Skarina, T., Mesa, N., Endres, M., Savchenko, A., Joachimiak, A., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Uncharacterized oxidoreductase YohF | 254 | Escherichia coli K-12 | Mutation(s): 0 Gene Names: yohF, yohE, b2137, JW2125 EC: 1 | 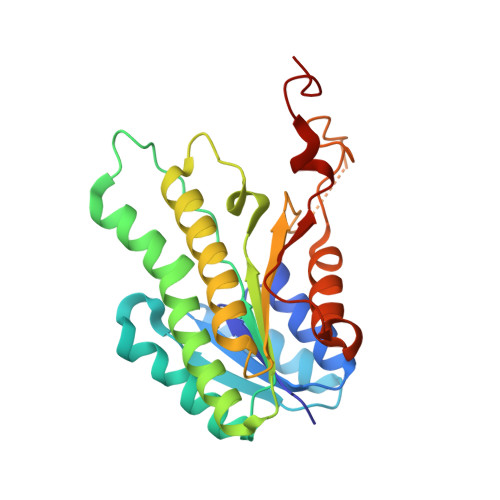 | |
UniProt | |||||
Find proteins for P33368 (Escherichia coli (strain K12)) Explore P33368 Go to UniProtKB: P33368 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P33368 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PGE Query on PGE | L [auth C], N [auth D] | TRIETHYLENE GLYCOL C6 H14 O4 ZIBGPFATKBEMQZ-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | P [auth E], Q [auth E], T [auth H] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| CA Query on CA | I [auth A] J [auth B] K [auth C] M [auth D] O [auth E] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 54.819 | α = 82.59 |
| b = 72.21 | β = 87.03 |
| c = 119.945 | γ = 67.82 |
| Software Name | Purpose |
|---|---|
| HKL-3000 | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| HKL-3000 | data reduction |
| HKL-3000 | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | HHSN272201200026C |
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | HHSN272201700060C |