Glycosylation Associate Protein (Gap123) complex from Streptococcus agalactiae
zhang, H., Zhu, F., Wu, H.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Currently 6OYC does not have a validation slider image.
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Glycosylation Associate Protein 1 | 514 | Streptococcus agalactiae 2603V/R | Mutation(s): 0 Gene Names: SAG1452 | 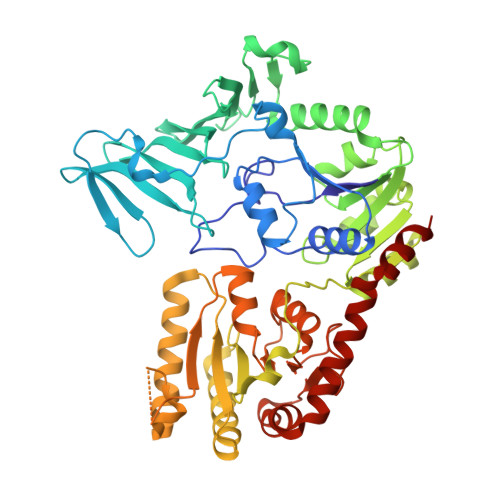 | |
UniProt | |||||
Find proteins for Q8DYM4 (Streptococcus agalactiae serotype V (strain ATCC BAA-611 / 2603 V/R)) Explore Q8DYM4 Go to UniProtKB: Q8DYM4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8DYM4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Glycosylation Associate Protein 2 | 519 | Streptococcus agalactiae 2603V/R | Mutation(s): 0 Gene Names: SAG1451 | 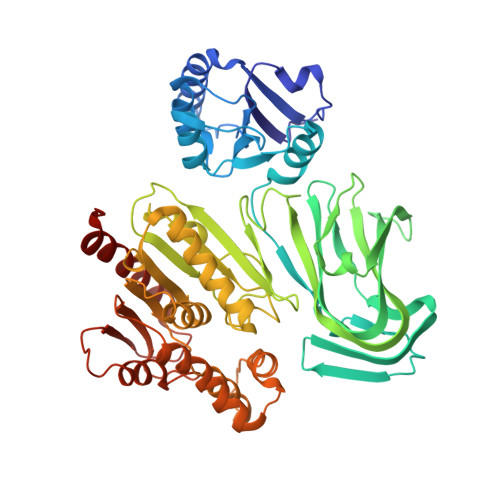 | |
UniProt | |||||
Find proteins for Q8DYM5 (Streptococcus agalactiae serotype V (strain ATCC BAA-611 / 2603 V/R)) Explore Q8DYM5 Go to UniProtKB: Q8DYM5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8DYM5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Glycosylation Associate Protein 3 | 330 | Streptococcus agalactiae 2603V/R | Mutation(s): 0 Gene Names: SAG1450 | 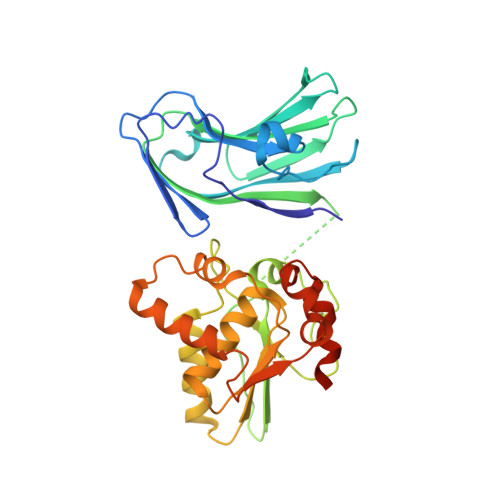 | |
UniProt | |||||
Find proteins for Q8DYM6 (Streptococcus agalactiae serotype V (strain ATCC BAA-611 / 2603 V/R)) Explore Q8DYM6 Go to UniProtKB: Q8DYM6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8DYM6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SO4 Query on SO4 | J [auth A], K [auth A], P [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| GOL Query on GOL | D [auth A] E [auth A] F [auth A] G [auth A] H [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 103.009 | α = 90 |
| b = 168.321 | β = 90 |
| c = 100.034 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHENIX | phasing |
Currently 6OYC does not have a validation slider image.
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of Dental and Craniofacial Research (NIH/NIDCR) | United States | 5R01DE017954-12 |